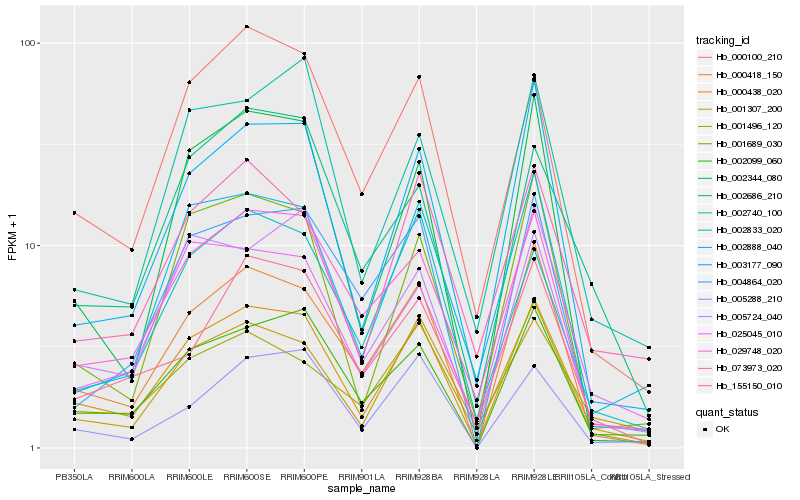
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000438_020 |
0.0 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 2 |
Hb_002099_060 |
0.0916270346 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 3 |
Hb_000418_150 |
0.0929950309 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_029748_020 |
0.11256622 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_003177_090 |
0.1153843517 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 6 |
Hb_001307_200 |
0.1182636407 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 7 |
Hb_001689_030 |
0.12040396 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 8 |
Hb_002888_040 |
0.1209920692 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 9 |
Hb_000100_210 |
0.1231591141 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 10 |
Hb_073973_020 |
0.1274960885 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 11 |
Hb_002344_080 |
0.1282496787 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 12 |
Hb_001496_120 |
0.1292948104 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_004864_020 |
0.12954217 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_005288_210 |
0.1306797427 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 15 |
Hb_002833_020 |
0.1307124812 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_005724_040 |
0.1307585548 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 17 |
Hb_025045_010 |
0.1335416435 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_002740_100 |
0.1352761925 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 19 |
Hb_002686_210 |
0.1363319067 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_155150_010 |
0.1386991574 |
- |
- |
kinase, putative [Ricinus communis] |