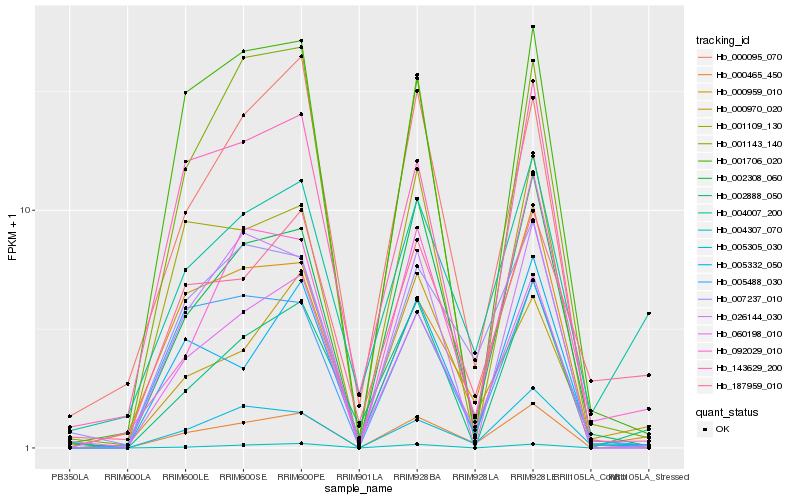
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_450 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_060198_010 |
0.1216935594 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_005305_030 |
0.1326006161 |
- |
- |
hypothetical protein JCGZ_25922 [Jatropha curcas] |
| 4 |
Hb_002308_060 |
0.1328955429 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 5 |
Hb_007237_010 |
0.1356151247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000959_010 |
0.1383805072 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004007_200 |
0.1485970035 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 8 |
Hb_092029_010 |
0.1500655917 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 9 |
Hb_005332_050 |
0.1524884364 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 10 |
Hb_001706_020 |
0.161567271 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 11 |
Hb_000970_020 |
0.161691202 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 12 |
Hb_001109_130 |
0.1627199653 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005488_030 |
0.1629520936 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 14 |
Hb_187959_010 |
0.1636435626 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 15 |
Hb_143629_200 |
0.1640237065 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 16 |
Hb_002888_050 |
0.1652077837 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001143_140 |
0.1671061139 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_026144_030 |
0.1671554981 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 19 |
Hb_004307_070 |
0.1703274016 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_000095_070 |
0.1716505153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |