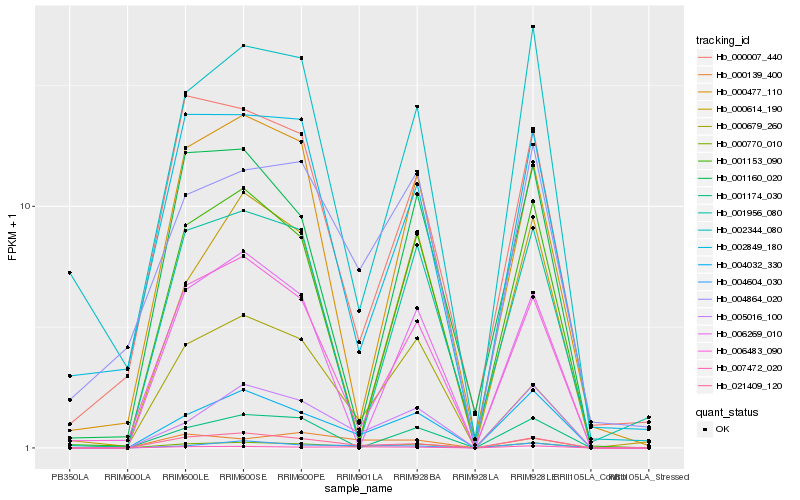
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000770_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 2 |
Hb_004032_330 |
0.0768258575 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005016_100 |
0.1148608022 |
- |
- |
- |
| 4 |
Hb_021409_120 |
0.1237363255 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 5 |
Hb_006483_090 |
0.1483923925 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 6 |
Hb_004864_020 |
0.152576843 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 7 |
Hb_000679_260 |
0.1597041516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001153_090 |
0.1615813286 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 9 |
Hb_007472_020 |
0.1617487836 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 10 |
Hb_000007_440 |
0.1680030806 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 11 |
Hb_002849_180 |
0.1740617487 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 12 |
Hb_000477_110 |
0.1741991927 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 13 |
Hb_004604_030 |
0.174247308 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_002344_080 |
0.175167375 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 15 |
Hb_000139_400 |
0.1769346641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006269_010 |
0.1797699634 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 17 |
Hb_001174_030 |
0.1813628792 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001956_080 |
0.182242447 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 19 |
Hb_000614_190 |
0.1827704116 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 20 |
Hb_001160_020 |
0.1835817197 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |