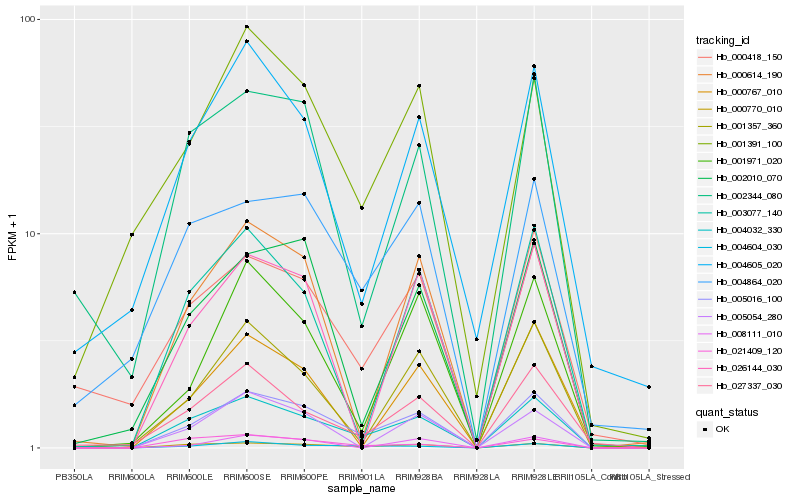
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005016_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_004032_330 |
0.0699432364 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000770_010 |
0.1148608022 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 4 |
Hb_004604_030 |
0.1423091408 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_002010_070 |
0.1514196327 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 6 |
Hb_000614_190 |
0.1577341285 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 7 |
Hb_001357_360 |
0.1590989461 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_027337_030 |
0.1593754728 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 9 |
Hb_001971_020 |
0.1603275055 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 10 |
Hb_001391_100 |
0.1645409718 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 11 |
Hb_000767_010 |
0.166365756 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 12 |
Hb_026144_030 |
0.1665496434 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 13 |
Hb_003077_140 |
0.1702180649 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 14 |
Hb_002344_080 |
0.1707898865 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 15 |
Hb_021409_120 |
0.1753329955 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_005054_280 |
0.1755267163 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 17 |
Hb_004864_020 |
0.1757189458 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 18 |
Hb_000418_150 |
0.1767478039 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_008111_010 |
0.1778118643 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 20 |
Hb_004605_020 |
0.1780965241 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |