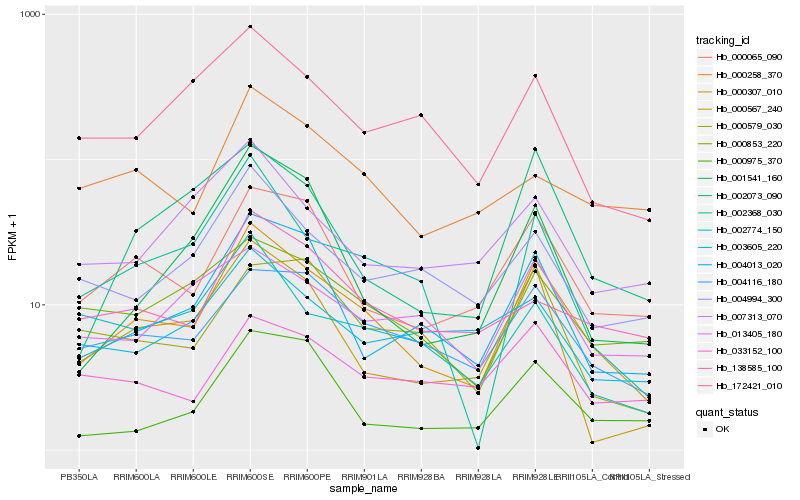
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_010 |
0.0 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 2 |
Hb_000065_090 |
0.1462937191 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_004116_180 |
0.1515572054 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 4 |
Hb_004013_020 |
0.1538826893 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 5 |
Hb_002368_030 |
0.1598315679 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 6 |
Hb_000567_240 |
0.1701323391 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 7 |
Hb_004994_300 |
0.1714944989 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 8 |
Hb_172421_010 |
0.1747093475 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 9 |
Hb_000853_220 |
0.1757249697 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 10 |
Hb_002073_090 |
0.1789541866 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |
| 11 |
Hb_138585_100 |
0.1821239704 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 12 |
Hb_000579_030 |
0.1836648432 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 13 |
Hb_001541_160 |
0.1841496879 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 14 |
Hb_013405_180 |
0.1863252566 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 15 |
Hb_007313_070 |
0.1876641919 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 16 |
Hb_000975_370 |
0.1879595955 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 17 |
Hb_033152_100 |
0.1885347858 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000258_370 |
0.1899191798 |
- |
- |
hypothetical protein [Ricinus communis] |
| 19 |
Hb_002774_150 |
0.1915591563 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 20 |
Hb_003605_220 |
0.1936951008 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |