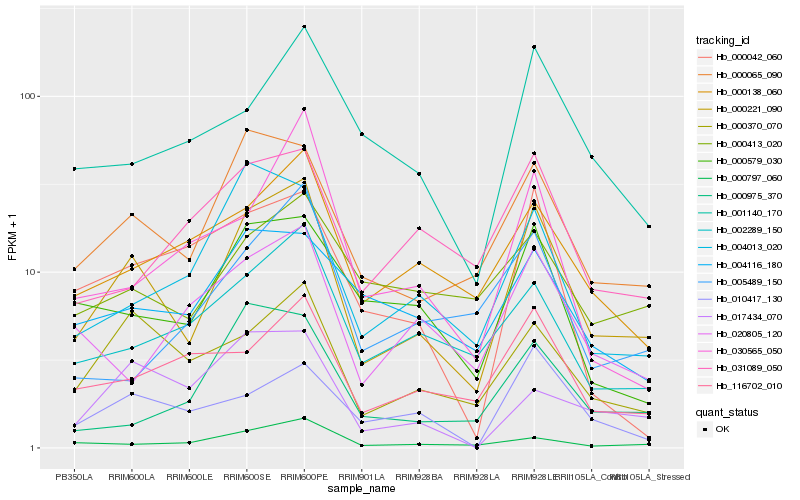
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000221_090 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 2 |
Hb_000065_090 |
0.1490242152 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000370_070 |
0.1791586838 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 4 |
Hb_004013_020 |
0.1946073345 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 5 |
Hb_000797_060 |
0.1951085164 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 6 |
Hb_002289_150 |
0.1973477315 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 7 |
Hb_000579_030 |
0.1977044701 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 8 |
Hb_000138_060 |
0.2002313217 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 9 |
Hb_116702_010 |
0.2048060928 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 10 |
Hb_000975_370 |
0.2086379537 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001140_170 |
0.2089057979 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 12 |
Hb_010417_130 |
0.2092344754 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 13 |
Hb_004116_180 |
0.2143257306 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 14 |
Hb_000413_020 |
0.2163535307 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 15 |
Hb_031089_050 |
0.2168915197 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_017434_070 |
0.2174864486 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_020805_120 |
0.2205105375 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 18 |
Hb_005489_150 |
0.2258453361 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 19 |
Hb_030565_050 |
0.2269859861 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 20 |
Hb_000042_060 |
0.2270052954 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |