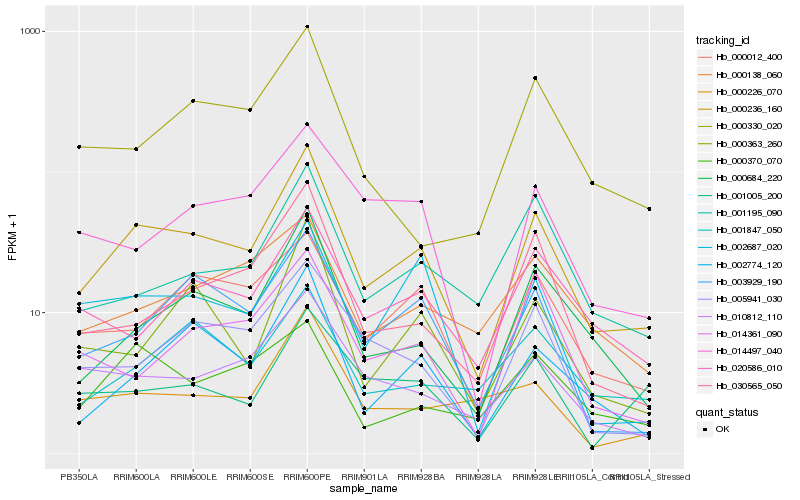
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_160 |
0.0 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 2 |
Hb_000684_220 |
0.1458782908 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002774_120 |
0.1543559064 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 4 |
Hb_003929_190 |
0.15688635 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 5 |
Hb_000012_400 |
0.1634705632 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 6 |
Hb_010812_110 |
0.1656800474 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 7 |
Hb_030565_050 |
0.1681976932 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 8 |
Hb_005941_030 |
0.169168711 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_020586_010 |
0.1719968364 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000363_260 |
0.1730119025 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 11 |
Hb_014497_040 |
0.1756869303 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001195_090 |
0.1795654368 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 13 |
Hb_001847_050 |
0.1802436569 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 14 |
Hb_000138_060 |
0.1825113464 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 15 |
Hb_002687_020 |
0.1837294566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000226_070 |
0.1850977203 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 17 |
Hb_000370_070 |
0.1859623939 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 18 |
Hb_014361_090 |
0.1864802812 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 19 |
Hb_000330_020 |
0.1881515961 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 20 |
Hb_001005_200 |
0.1896756235 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |