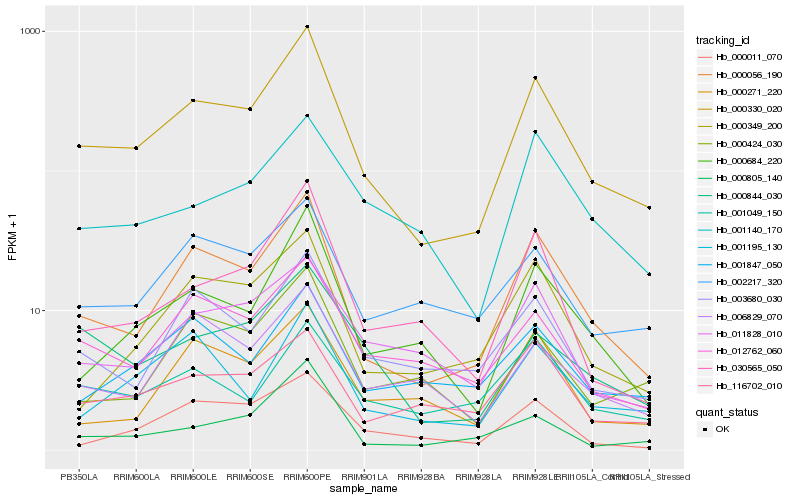
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000330_020 |
0.0 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 2 |
Hb_000056_190 |
0.0870991445 |
- |
- |
hypothetical protein [Ricinus communis] |
| 3 |
Hb_000684_220 |
0.143023375 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_030565_050 |
0.1440697954 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 5 |
Hb_012762_060 |
0.149201577 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_003680_030 |
0.1506977436 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000844_030 |
0.1551861166 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 8 |
Hb_000349_200 |
0.1589465791 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 9 |
Hb_011828_010 |
0.1590962402 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 10 |
Hb_002217_320 |
0.1595111504 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_116702_010 |
0.1620127183 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 12 |
Hb_000424_030 |
0.1634963504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000805_140 |
0.164303347 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 14 |
Hb_001195_130 |
0.1661506863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000271_220 |
0.1682896259 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_001847_050 |
0.1684044246 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 17 |
Hb_006829_070 |
0.1684616098 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 18 |
Hb_000011_070 |
0.1696705277 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 19 |
Hb_001049_150 |
0.1702478477 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 20 |
Hb_001140_170 |
0.1704004969 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |