| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
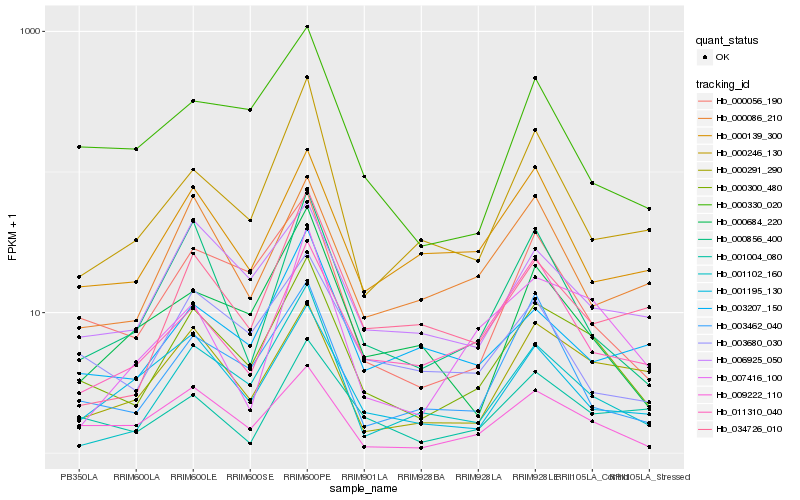
Hb_000300_480 |
0.0 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 2 |
Hb_009222_110 |
0.1348769865 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 3 |
Hb_000056_190 |
0.1368821364 |
- |
- |
hypothetical protein [Ricinus communis] |
| 4 |
Hb_006925_050 |
0.161440692 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 5 |
Hb_001102_160 |
0.1653179529 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 6 |
Hb_000291_290 |
0.1688208973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000856_400 |
0.1697654237 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 8 |
Hb_003680_030 |
0.1733617829 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000330_020 |
0.1764562151 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 10 |
Hb_000246_130 |
0.1772880525 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 11 |
Hb_003462_040 |
0.1794401082 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_001004_080 |
0.1800960917 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 13 |
Hb_011310_040 |
0.1810923346 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 14 |
Hb_001195_130 |
0.181382677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007416_100 |
0.1838042817 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 16 |
Hb_000684_220 |
0.1846329797 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_003207_150 |
0.186099422 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 18 |
Hb_034726_010 |
0.1868032259 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_000139_300 |
0.1877729142 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 20 |
Hb_000086_210 |
0.1877931608 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |