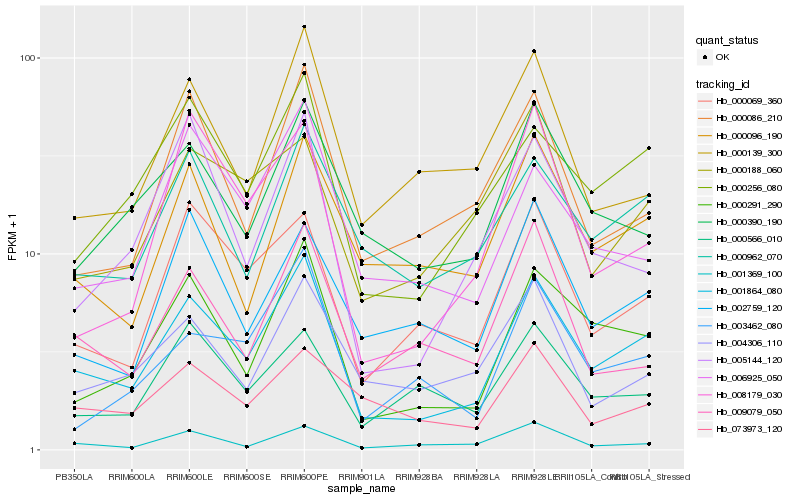
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001864_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 2 |
Hb_000291_290 |
0.1155993253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000256_080 |
0.1315836133 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 4 |
Hb_000962_070 |
0.1467647214 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 13, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_000069_360 |
0.1527103927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000390_190 |
0.1563038367 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000096_190 |
0.1574108749 |
- |
- |
unknown [Lotus japonicus] |
| 8 |
Hb_000086_210 |
0.1591973381 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_006925_050 |
0.1600924613 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 10 |
Hb_001369_100 |
0.1606399658 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 11 |
Hb_003462_080 |
0.1670077871 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000139_300 |
0.1674559011 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 13 |
Hb_009079_050 |
0.1683060643 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 14 |
Hb_004306_110 |
0.1698829098 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 15 |
Hb_002759_120 |
0.1701581511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005144_120 |
0.1708541216 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 17 |
Hb_073973_120 |
0.1728496282 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000566_010 |
0.1733682787 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 19 |
Hb_008179_030 |
0.1734175313 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 8 [Jatropha curcas] |
| 20 |
Hb_000188_060 |
0.1757486056 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |