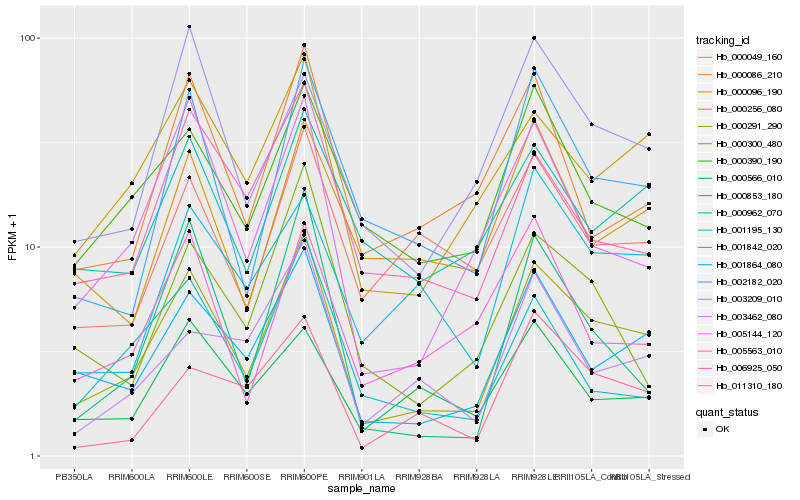
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_290 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001864_080 |
0.1155993253 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 3 |
Hb_000256_080 |
0.1400297459 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 4 |
Hb_002182_020 |
0.1433112823 |
- |
- |
Thylakoid lumenal 17.4 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_005144_120 |
0.1521963344 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 6 |
Hb_000390_190 |
0.1596797542 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_006925_050 |
0.1634616077 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 8 |
Hb_000962_070 |
0.1647818352 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 13, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_000086_210 |
0.1651276978 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_000300_480 |
0.1688208973 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 11 |
Hb_011310_180 |
0.1689580466 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003462_080 |
0.1705291353 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000096_190 |
0.1725462692 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_001842_020 |
0.1728048683 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 15 |
Hb_000049_160 |
0.1734310365 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 16 |
Hb_000853_180 |
0.1738736954 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 17 |
Hb_001195_130 |
0.1748157217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003209_010 |
0.1756265511 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 19 |
Hb_005563_010 |
0.1765919784 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 20 |
Hb_000566_010 |
0.1795106247 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |