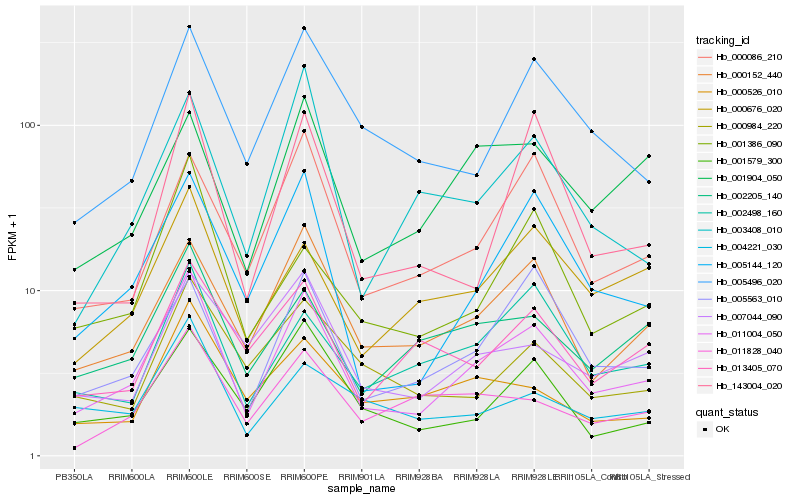
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011004_050 |
0.0 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 2 |
Hb_000152_440 |
0.1372071603 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 3 |
Hb_005144_120 |
0.1376322659 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 4 |
Hb_001579_300 |
0.1521787795 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 5 |
Hb_000086_210 |
0.1550401888 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_005563_010 |
0.157157336 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 7 |
Hb_005496_020 |
0.1608051797 |
- |
- |
PREDICTED: uncharacterized protein LOC105630867 [Jatropha curcas] |
| 8 |
Hb_002205_140 |
0.1611744112 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 9 |
Hb_002498_160 |
0.1626204732 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 10 |
Hb_000676_020 |
0.1628409732 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 11 |
Hb_007044_090 |
0.166978606 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 12 |
Hb_011828_040 |
0.1684125605 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001904_050 |
0.1686344487 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 14 |
Hb_001386_090 |
0.1719590718 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 15 |
Hb_013405_070 |
0.1723872563 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 16 |
Hb_003408_010 |
0.1733764544 |
- |
- |
fructokinase [Manihot esculenta] |
| 17 |
Hb_143004_020 |
0.1742866356 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 18 |
Hb_000526_010 |
0.1745531945 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 19 |
Hb_004221_030 |
0.1747261206 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000984_220 |
0.1750518098 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |