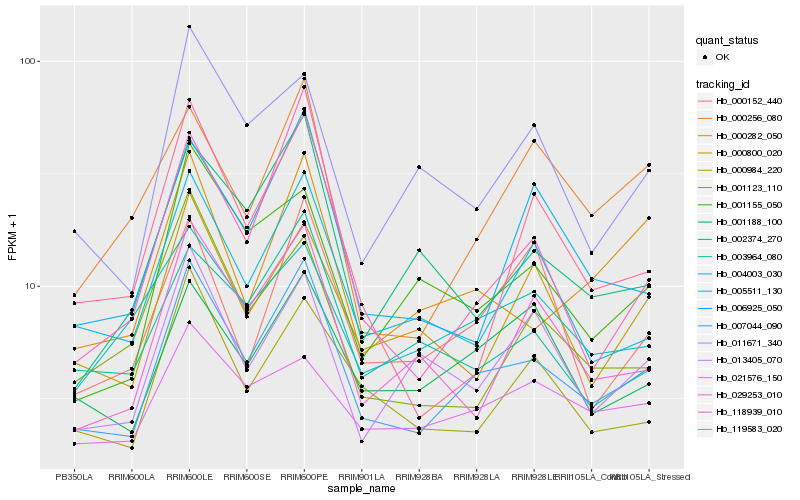
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_090 |
0.0 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 2 |
Hb_005511_130 |
0.1273232702 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 3 |
Hb_000984_220 |
0.1335725828 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 4 |
Hb_029253_010 |
0.1365536621 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 5 |
Hb_006925_050 |
0.1387355594 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 6 |
Hb_003964_080 |
0.1392750802 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 7 |
Hb_000282_050 |
0.1415844646 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 8 |
Hb_001188_100 |
0.1422319829 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 9 |
Hb_118939_010 |
0.1426294831 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_000800_020 |
0.1427514979 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 11 |
Hb_001155_050 |
0.1430395832 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 12 |
Hb_002374_270 |
0.1453742146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011671_340 |
0.1464260376 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 14 |
Hb_013405_070 |
0.146661883 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 15 |
Hb_001123_110 |
0.1501266314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000152_440 |
0.1529185381 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 17 |
Hb_004003_030 |
0.1533746183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_021576_150 |
0.1546525343 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 19 |
Hb_119583_020 |
0.1547000244 |
- |
- |
PREDICTED: uncharacterized protein LOC105628090 [Jatropha curcas] |
| 20 |
Hb_000256_080 |
0.1549932718 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |