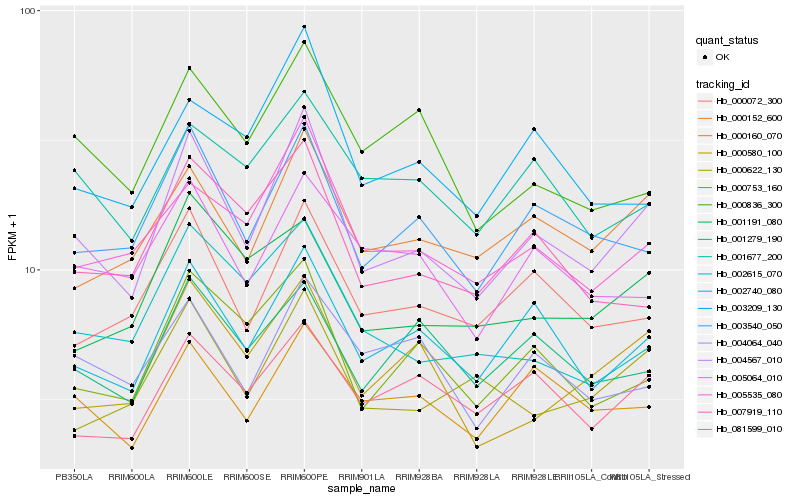
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004567_010 |
0.0 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 2 |
Hb_002740_080 |
0.0922978429 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 3 |
Hb_003540_050 |
0.1014968901 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 4 |
Hb_000152_600 |
0.1085287494 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 5 |
Hb_005535_080 |
0.1092433907 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 6 |
Hb_000072_300 |
0.1120283973 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 7 |
Hb_000753_160 |
0.1138607605 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 8 |
Hb_000160_070 |
0.1141974011 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 9 |
Hb_000622_130 |
0.1194142979 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 10 |
Hb_081599_010 |
0.121415421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000580_100 |
0.1214760103 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003209_130 |
0.1244720485 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 13 |
Hb_007919_110 |
0.1260004912 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 14 |
Hb_001279_190 |
0.1268782186 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005064_010 |
0.1271463944 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 16 |
Hb_001191_080 |
0.1274443573 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_002615_070 |
0.1292942706 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004064_040 |
0.1293825913 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 19 |
Hb_001677_200 |
0.1294915992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000836_300 |
0.1302655358 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |