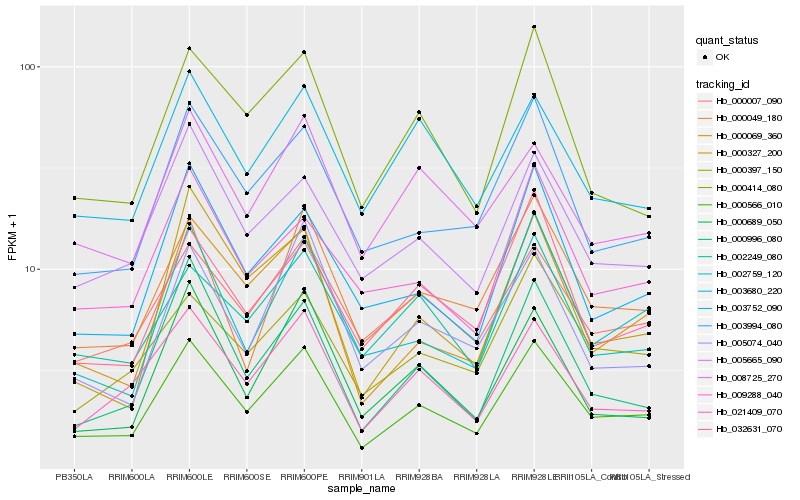
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000566_010 |
0.0 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 2 |
Hb_000069_360 |
0.0870389671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003994_080 |
0.0923000484 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 4 |
Hb_021409_070 |
0.0931853313 |
- |
- |
PREDICTED: transcription factor bHLH157 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002759_120 |
0.095761533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003752_090 |
0.1011378521 |
- |
- |
chitinase, putative [Ricinus communis] |
| 7 |
Hb_005665_090 |
0.1020516008 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 8 |
Hb_000414_080 |
0.1041647382 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000996_080 |
0.1066023639 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 10 |
Hb_000007_090 |
0.106843606 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 11 |
Hb_032631_070 |
0.1082316753 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_000397_150 |
0.1106910759 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 13 |
Hb_002249_080 |
0.1111368188 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 14 |
Hb_000049_180 |
0.1113487872 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 15 |
Hb_003680_220 |
0.111885821 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 16 |
Hb_009288_040 |
0.1122706027 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 17 |
Hb_005074_040 |
0.1136677462 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_008725_270 |
0.1137523199 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 19 |
Hb_000327_200 |
0.1145099593 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 20 |
Hb_000689_050 |
0.1146963335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |