| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
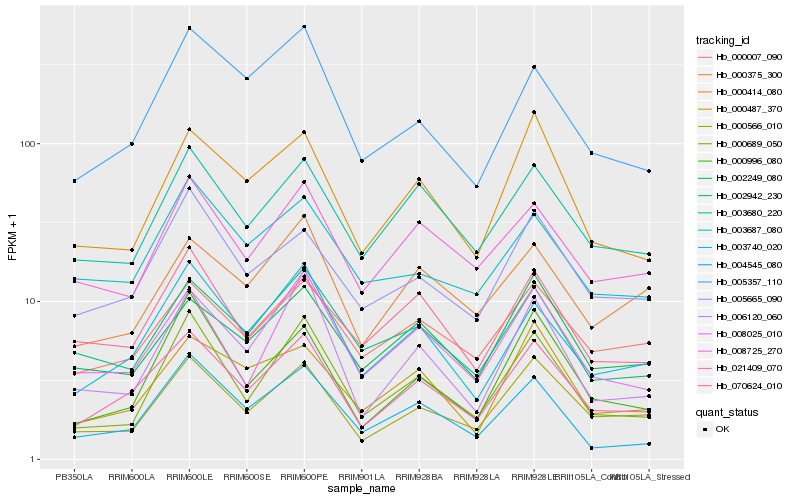
Hb_021409_070 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH157 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003740_020 |
0.0894892503 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_000566_010 |
0.0931853313 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 4 |
Hb_003680_220 |
0.1042146015 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 5 |
Hb_000007_090 |
0.1061139758 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 6 |
Hb_005665_090 |
0.106591596 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 7 |
Hb_000414_080 |
0.1101065781 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008725_270 |
0.1131156047 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 9 |
Hb_008025_010 |
0.116380034 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 10 |
Hb_005357_110 |
0.1168168868 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 11 |
Hb_006120_060 |
0.116876521 |
- |
- |
O-acetyltransferase family protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_000689_050 |
0.1187678932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002942_230 |
0.1200271686 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 14 |
Hb_004545_080 |
0.1203999175 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_002249_080 |
0.1205226919 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 16 |
Hb_000487_370 |
0.121716463 |
- |
- |
- |
| 17 |
Hb_000375_300 |
0.1221704718 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 18 |
Hb_000996_080 |
0.1229995902 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 19 |
Hb_070624_010 |
0.1241148639 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 20 |
Hb_003687_080 |
0.1247421035 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |