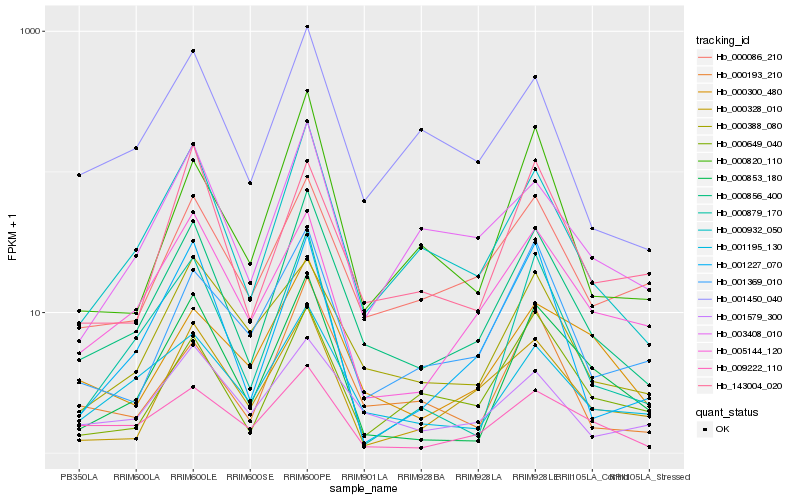
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_400 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 2 |
Hb_000932_050 |
0.1002707209 |
- |
- |
annexin [Manihot esculenta] |
| 3 |
Hb_001195_130 |
0.1403944768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000853_180 |
0.1422963171 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 5 |
Hb_001450_040 |
0.1458795029 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 6 |
Hb_000193_210 |
0.1458843143 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 7 |
Hb_001579_300 |
0.1521987605 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 8 |
Hb_000086_210 |
0.1543337096 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_003408_010 |
0.155123654 |
- |
- |
fructokinase [Manihot esculenta] |
| 10 |
Hb_000879_170 |
0.1565883781 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000820_110 |
0.1592475901 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 12 |
Hb_005144_120 |
0.1601077839 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 13 |
Hb_000328_010 |
0.1606132025 |
- |
- |
PREDICTED: QWRF motif-containing protein 8-like [Jatropha curcas] |
| 14 |
Hb_000388_080 |
0.1612220695 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 15 |
Hb_001369_010 |
0.1638459303 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 16 |
Hb_001227_070 |
0.1643770615 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 17 |
Hb_009222_110 |
0.1657620882 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 18 |
Hb_000649_040 |
0.1659077568 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 19 |
Hb_143004_020 |
0.1683025348 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 20 |
Hb_000300_480 |
0.1697654237 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |