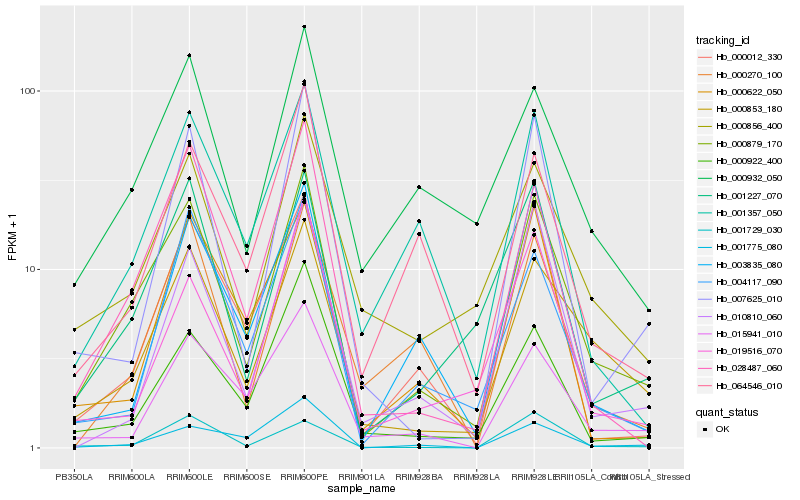
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000879_170 |
0.0 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000853_180 |
0.1265915053 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 3 |
Hb_001227_070 |
0.1396862038 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 4 |
Hb_028487_060 |
0.1471672991 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 5 |
Hb_000856_400 |
0.1565883781 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 6 |
Hb_000012_330 |
0.1614135473 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 7 |
Hb_001357_050 |
0.1672810585 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 8 |
Hb_000932_050 |
0.1680646007 |
- |
- |
annexin [Manihot esculenta] |
| 9 |
Hb_015941_010 |
0.1729083182 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 10 |
Hb_001729_030 |
0.1730587296 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |
| 11 |
Hb_007625_010 |
0.1739114525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000270_100 |
0.1748748729 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 13 |
Hb_000922_400 |
0.1751090321 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 14 |
Hb_010810_060 |
0.1773925406 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 15 |
Hb_004117_090 |
0.1781573345 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 16 |
Hb_064546_010 |
0.1795784784 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 17 |
Hb_019516_070 |
0.1795827222 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 18 |
Hb_003835_080 |
0.1815020231 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000622_050 |
0.1822301325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001775_080 |
0.1835923837 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |