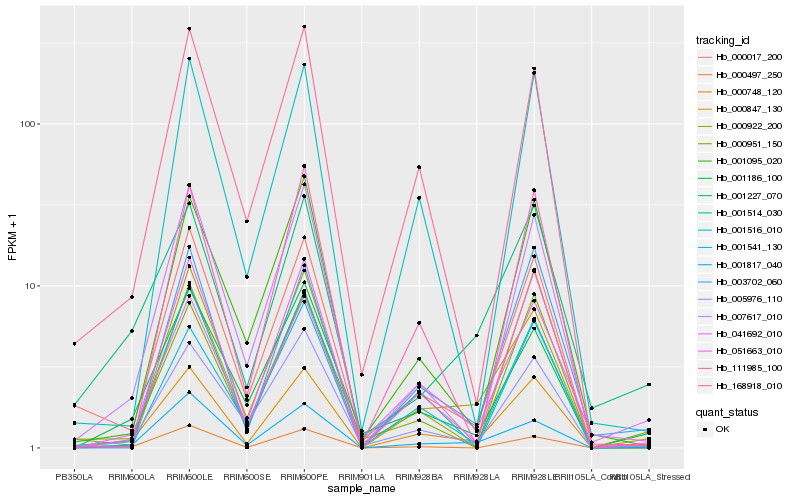
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000748_120 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 2 |
Hb_051663_010 |
0.0994613369 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_041692_010 |
0.1009744035 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_000847_130 |
0.1244680144 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 5 |
Hb_005976_110 |
0.1257588227 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 6 |
Hb_007617_010 |
0.1260883803 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001817_040 |
0.1306746519 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 8 |
Hb_003702_060 |
0.1337195778 |
- |
- |
PREDICTED: protein trichome birefringence-like 38 [Jatropha curcas] |
| 9 |
Hb_000017_200 |
0.1366120129 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 10 |
Hb_001516_010 |
0.1371808387 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 11 |
Hb_001186_100 |
0.137656106 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001541_130 |
0.1380718684 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 13 |
Hb_000951_150 |
0.1418968769 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 14 |
Hb_000922_200 |
0.1433102734 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 15 |
Hb_001514_030 |
0.1440296311 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 16 |
Hb_168918_010 |
0.1451137513 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 17 |
Hb_001227_070 |
0.1458392957 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 18 |
Hb_000497_250 |
0.1458420536 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 19 |
Hb_111985_100 |
0.148464445 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 20 |
Hb_001095_020 |
0.1486506446 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |