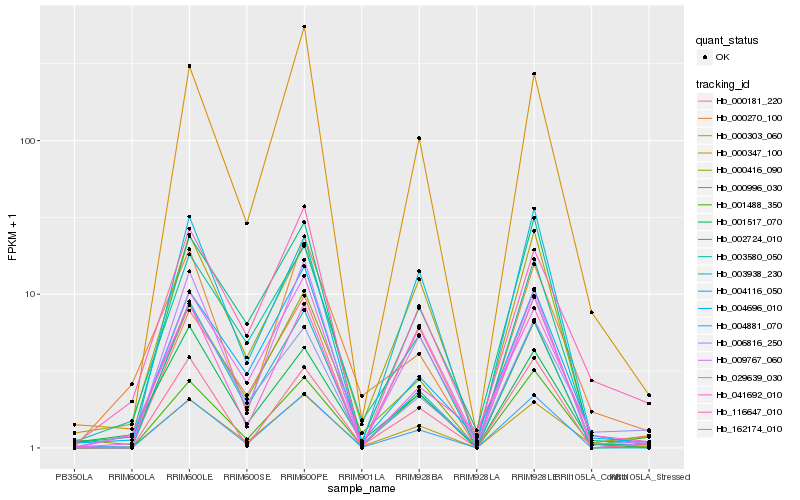
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_100 |
0.0 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 2 |
Hb_009767_060 |
0.1156057045 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 3 |
Hb_003580_050 |
0.1197656504 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 4 |
Hb_116647_010 |
0.121716731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_041692_010 |
0.1217429627 |
- |
- |
annexin [Manihot esculenta] |
| 6 |
Hb_000416_090 |
0.12269497 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 7 |
Hb_029639_030 |
0.126348743 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 8 |
Hb_004881_070 |
0.1270206229 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000303_060 |
0.1274704122 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000181_220 |
0.1301504981 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000996_030 |
0.1343145792 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004696_010 |
0.137150801 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 13 |
Hb_000270_100 |
0.1382587932 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 14 |
Hb_001488_350 |
0.1392114584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001517_070 |
0.139511257 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_006816_250 |
0.141554072 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 17 |
Hb_003938_230 |
0.1437766677 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 18 |
Hb_002724_010 |
0.1438584367 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 19 |
Hb_162174_010 |
0.1452005141 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_004116_050 |
0.1456989096 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |