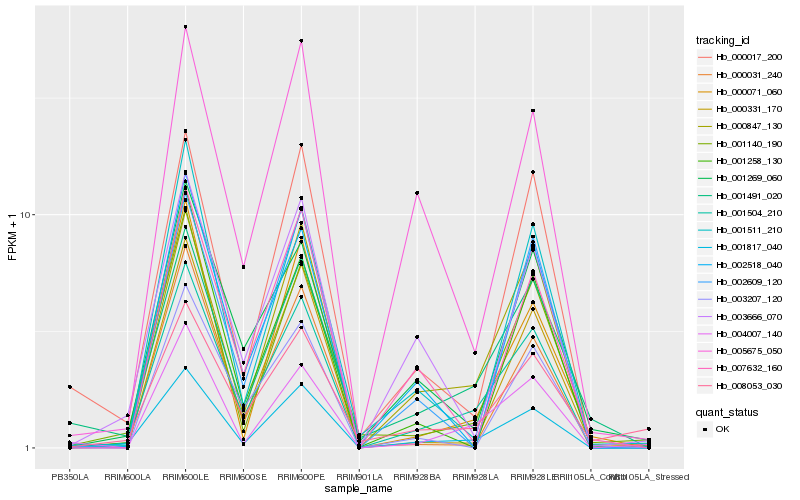
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001504_210 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001817_040 |
0.0927692342 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 3 |
Hb_000847_130 |
0.0982194897 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 4 |
Hb_004007_140 |
0.1386717761 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001140_190 |
0.1396931 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 6 |
Hb_001491_020 |
0.1410766504 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000071_060 |
0.1421656461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000331_170 |
0.1450421092 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 9 |
Hb_001269_060 |
0.1508711638 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001511_210 |
0.1532326113 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 11 |
Hb_000017_200 |
0.1542844443 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 12 |
Hb_001258_130 |
0.1554347913 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 13 |
Hb_000031_240 |
0.1559329494 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 14 |
Hb_002518_040 |
0.1560833345 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 15 |
Hb_005675_050 |
0.1569549575 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 16 |
Hb_003666_070 |
0.1577347744 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 17 |
Hb_002609_120 |
0.1591543498 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 18 |
Hb_007632_160 |
0.1592161597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008053_030 |
0.1592867653 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 20 |
Hb_003207_120 |
0.1609355667 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |