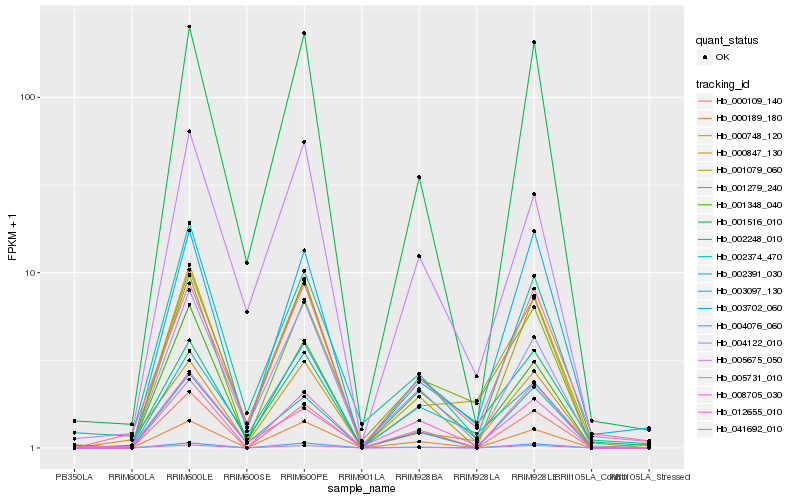
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_240 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105633500 [Jatropha curcas] |
| 2 |
Hb_000847_130 |
0.1133725809 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 3 |
Hb_001348_040 |
0.1291731506 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 4 |
Hb_004122_010 |
0.1403229486 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 5 |
Hb_003097_130 |
0.1469448036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000109_140 |
0.1510480856 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 7 |
Hb_041692_010 |
0.1550393582 |
- |
- |
annexin [Manihot esculenta] |
| 8 |
Hb_005731_010 |
0.1571955736 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 9 |
Hb_005675_050 |
0.1589367796 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 10 |
Hb_000189_180 |
0.1594591519 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 11 |
Hb_001516_010 |
0.1603851976 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 12 |
Hb_004076_060 |
0.1624453658 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 13 |
Hb_002391_030 |
0.1629303772 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 14 |
Hb_003702_060 |
0.1632726079 |
- |
- |
PREDICTED: protein trichome birefringence-like 38 [Jatropha curcas] |
| 15 |
Hb_012655_010 |
0.1634192411 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 16 |
Hb_000748_120 |
0.1634474687 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 17 |
Hb_002374_470 |
0.1640185167 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_002248_010 |
0.1648127484 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 19 |
Hb_001079_060 |
0.1667386849 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008705_030 |
0.1667543904 |
- |
- |
hypothetical protein JCGZ_13017 [Jatropha curcas] |