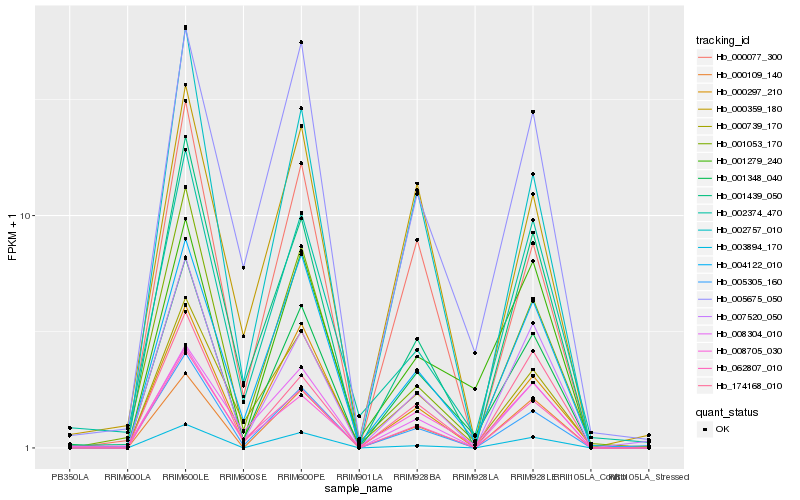
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_040 |
0.0 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 2 |
Hb_004122_010 |
0.1097150685 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 3 |
Hb_062807_010 |
0.1113165861 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 4 |
Hb_002757_010 |
0.1156076434 |
- |
- |
hypothetical protein POPTR_0013s10580g [Populus trichocarpa] |
| 5 |
Hb_005305_160 |
0.1191125856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000077_300 |
0.1212660301 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 7 |
Hb_000109_140 |
0.1226476308 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 8 |
Hb_000739_170 |
0.1236063681 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 9 |
Hb_005675_050 |
0.1238372895 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 10 |
Hb_008705_030 |
0.1272206889 |
- |
- |
hypothetical protein JCGZ_13017 [Jatropha curcas] |
| 11 |
Hb_001279_240 |
0.1291731506 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105633500 [Jatropha curcas] |
| 12 |
Hb_000297_210 |
0.1298550617 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 13 |
Hb_001439_050 |
0.1300101051 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_007520_050 |
0.1301464551 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 15 |
Hb_008304_010 |
0.133538435 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_000359_180 |
0.1341285524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003894_170 |
0.1387972312 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 18 |
Hb_174168_010 |
0.1403098435 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 19 |
Hb_001053_170 |
0.1404917288 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 20 |
Hb_002374_470 |
0.1441939188 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |