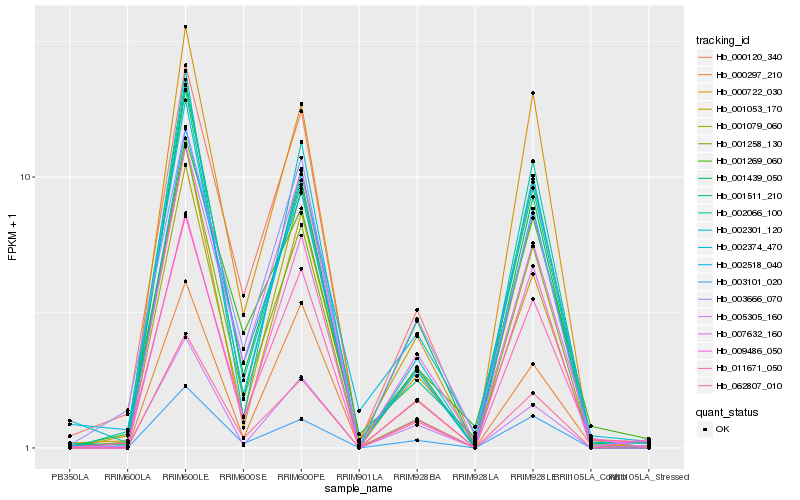
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_050 |
0.0 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 2 |
Hb_000120_340 |
0.08791752 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 3 |
Hb_002518_040 |
0.0884657069 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 4 |
Hb_001439_050 |
0.0931591408 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_001511_210 |
0.096561417 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 6 |
Hb_003666_070 |
0.0999501487 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 7 |
Hb_007632_160 |
0.1007077584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001258_130 |
0.1009485523 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 9 |
Hb_002374_470 |
0.1042615216 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_001079_060 |
0.1052718742 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002066_100 |
0.105855906 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 12 |
Hb_000722_030 |
0.1066430474 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 13 |
Hb_002301_120 |
0.1079769788 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 14 |
Hb_001053_170 |
0.1093221834 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 15 |
Hb_062807_010 |
0.1093235327 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 16 |
Hb_001269_060 |
0.1098183491 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003101_020 |
0.110025184 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 18 |
Hb_000297_210 |
0.1117329269 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 19 |
Hb_009486_050 |
0.1130957158 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 20 |
Hb_005305_160 |
0.1142445196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |