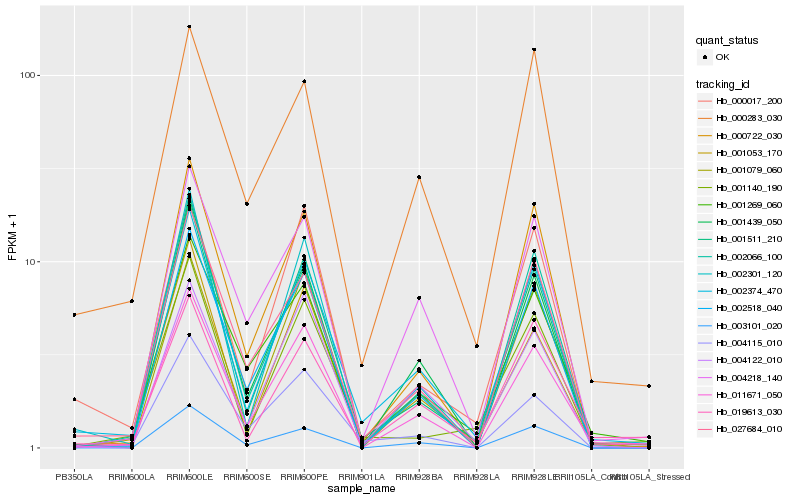
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_470 |
0.0 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_001511_210 |
0.088617691 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 3 |
Hb_011671_050 |
0.1042615216 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 4 |
Hb_027684_010 |
0.107605875 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 5 |
Hb_019613_030 |
0.1133143689 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 6 |
Hb_002066_100 |
0.1155236311 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 7 |
Hb_002518_040 |
0.1165812699 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 8 |
Hb_001053_170 |
0.1173727734 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 9 |
Hb_002301_120 |
0.11867861 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 10 |
Hb_001439_050 |
0.1190554761 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_000722_030 |
0.1191281389 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 12 |
Hb_001269_060 |
0.1223776793 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000283_030 |
0.1253606102 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 14 |
Hb_001140_190 |
0.1275213162 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 15 |
Hb_004115_010 |
0.1276943305 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 16 |
Hb_004218_140 |
0.1282724178 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 17 |
Hb_003101_020 |
0.1286204295 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 18 |
Hb_001079_060 |
0.1304797406 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004122_010 |
0.1306848845 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 20 |
Hb_000017_200 |
0.1309657065 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |