| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
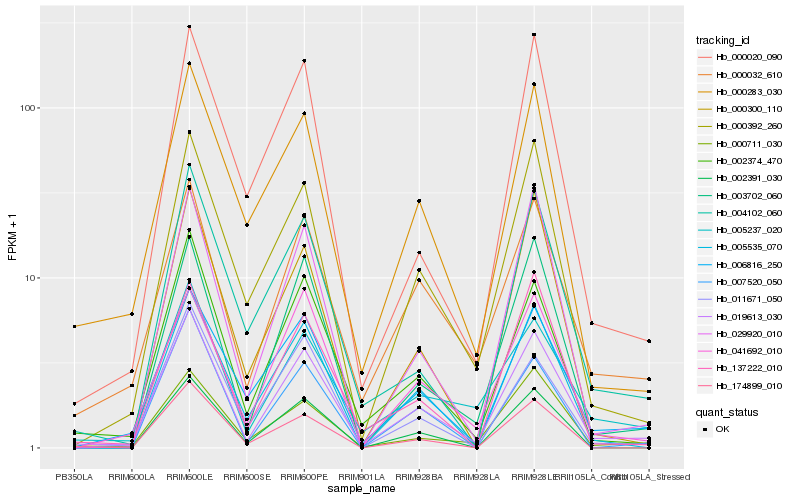
Hb_019613_030 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 2 |
Hb_002374_470 |
0.1133143689 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 3 |
Hb_000032_610 |
0.1133484393 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000392_260 |
0.1191004804 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 5 |
Hb_003702_060 |
0.1219501487 |
- |
- |
PREDICTED: protein trichome birefringence-like 38 [Jatropha curcas] |
| 6 |
Hb_000711_030 |
0.1225558299 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 7 |
Hb_029920_010 |
0.1255467227 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 8 |
Hb_006816_250 |
0.1269218075 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 9 |
Hb_000283_030 |
0.1278824296 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 10 |
Hb_005535_070 |
0.1295349984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005237_020 |
0.1323235901 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 12 |
Hb_041692_010 |
0.1360370405 |
- |
- |
annexin [Manihot esculenta] |
| 13 |
Hb_004102_060 |
0.1377272372 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 14 |
Hb_002391_030 |
0.1377649116 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 15 |
Hb_137222_010 |
0.1380812543 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 16 |
Hb_007520_050 |
0.1392266904 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 17 |
Hb_174899_010 |
0.1404954642 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 18 |
Hb_000020_090 |
0.1423796845 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 19 |
Hb_011671_050 |
0.1431622931 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 20 |
Hb_000300_110 |
0.1433826329 |
- |
- |
hypothetical protein JCGZ_02651 [Jatropha curcas] |