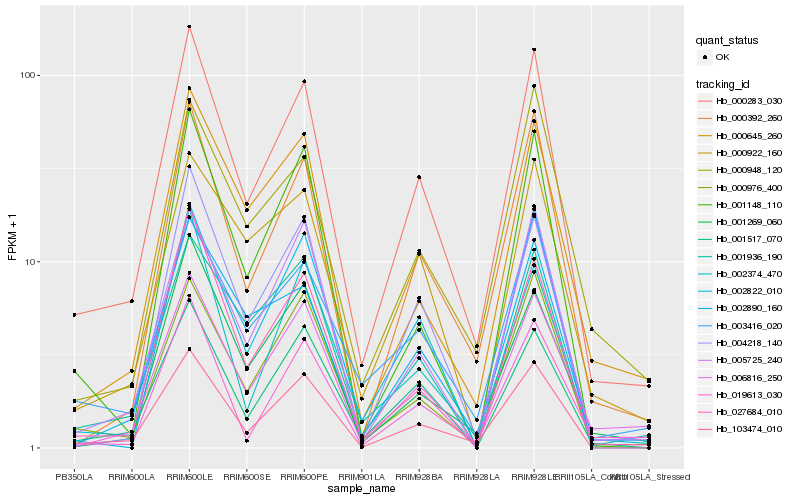
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000283_030 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 2 |
Hb_103474_010 |
0.0728738351 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000392_260 |
0.0937909614 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 4 |
Hb_000976_400 |
0.1040268774 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 5 |
Hb_006816_250 |
0.1076888019 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 6 |
Hb_003416_020 |
0.1102726191 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 7 |
Hb_001517_070 |
0.1105931059 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_005725_240 |
0.1130324931 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002890_160 |
0.1136446887 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004218_140 |
0.1140520889 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 11 |
Hb_001936_190 |
0.1191117623 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 12 |
Hb_027684_010 |
0.1220824454 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 13 |
Hb_000645_260 |
0.1238011574 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 14 |
Hb_002374_470 |
0.1253606102 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 15 |
Hb_002822_010 |
0.1259918461 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 16 |
Hb_001269_060 |
0.1262204545 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000948_120 |
0.1275770348 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 18 |
Hb_019613_030 |
0.1278824296 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 19 |
Hb_000922_160 |
0.1286541112 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 20 |
Hb_001148_110 |
0.1304802042 |
- |
- |
- |