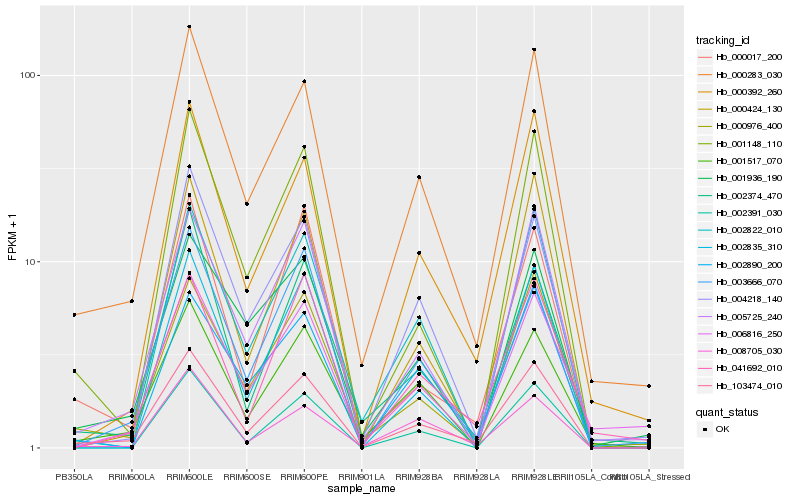
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103474_010 |
0.0 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000283_030 |
0.0728738351 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 3 |
Hb_000976_400 |
0.1093386499 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 4 |
Hb_000392_260 |
0.1120648461 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 5 |
Hb_005725_240 |
0.1121316612 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001517_070 |
0.1211928836 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_001148_110 |
0.1254819671 |
- |
- |
- |
| 8 |
Hb_000424_130 |
0.1285056172 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 9 |
Hb_000017_200 |
0.1311523745 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 10 |
Hb_002822_010 |
0.1317510678 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 11 |
Hb_002374_470 |
0.135514794 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_004218_140 |
0.1370880822 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 13 |
Hb_002890_200 |
0.1370889186 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 14 |
Hb_008705_030 |
0.1384736264 |
- |
- |
hypothetical protein JCGZ_13017 [Jatropha curcas] |
| 15 |
Hb_002835_310 |
0.1394518988 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_041692_010 |
0.1394960101 |
- |
- |
annexin [Manihot esculenta] |
| 17 |
Hb_006816_250 |
0.1402510216 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 18 |
Hb_002391_030 |
0.1402704769 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 19 |
Hb_001936_190 |
0.14050056 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 20 |
Hb_003666_070 |
0.1420443897 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |