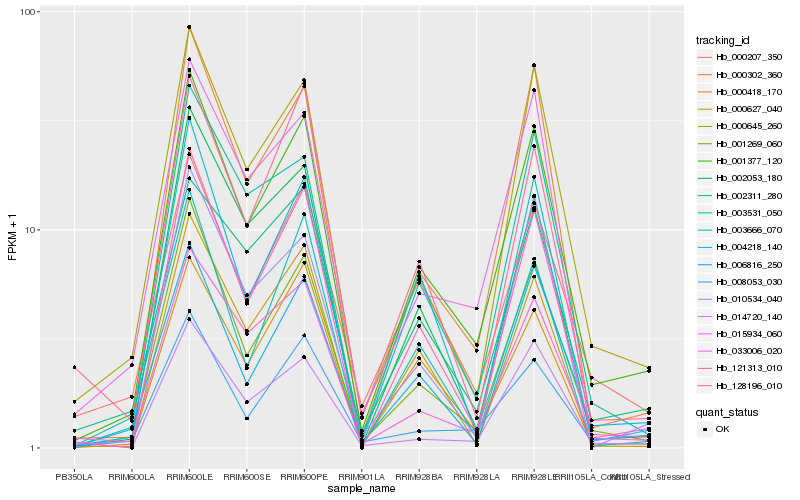
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 2 |
Hb_001269_060 |
0.06839178 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 3 |
Hb_121313_010 |
0.090876284 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 4 |
Hb_000645_260 |
0.0926755409 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 5 |
Hb_000302_360 |
0.0954889495 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_033006_020 |
0.0975093142 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 7 |
Hb_006816_250 |
0.1080868276 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 8 |
Hb_003531_050 |
0.1107670452 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_008053_030 |
0.1126470152 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 10 |
Hb_004218_140 |
0.1126896371 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000207_350 |
0.1171900854 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 12 |
Hb_015934_060 |
0.1185944262 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 13 |
Hb_002311_280 |
0.1213302053 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 14 |
Hb_000418_170 |
0.1224442489 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 15 |
Hb_010534_040 |
0.1233546002 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000627_040 |
0.1243675549 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 17 |
Hb_002053_180 |
0.1245071761 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 18 |
Hb_014720_140 |
0.1260321407 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 19 |
Hb_128196_010 |
0.1313723166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003666_070 |
0.1321005315 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |