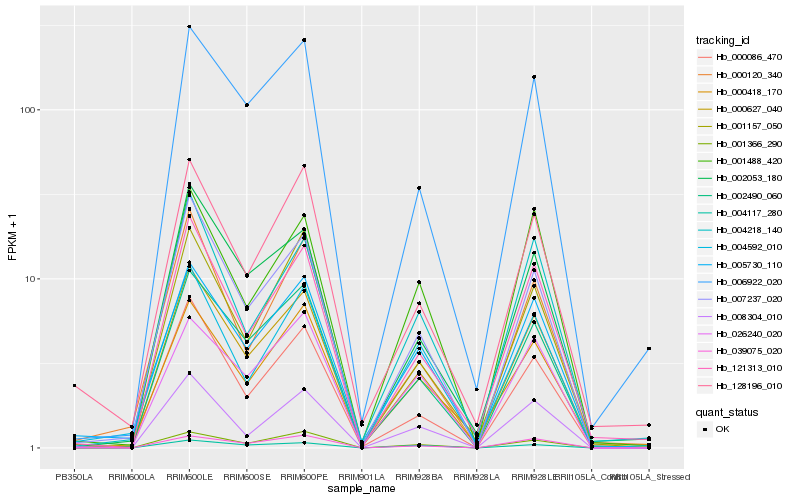
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000627_040 |
0.0 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 2 |
Hb_002490_060 |
0.0624290907 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 3 |
Hb_007237_020 |
0.0676722534 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 4 |
Hb_121313_010 |
0.0697197019 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 5 |
Hb_005730_110 |
0.0758374509 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 6 |
Hb_002053_180 |
0.0802277132 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 7 |
Hb_006922_020 |
0.0803875609 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 8 |
Hb_000086_470 |
0.0851224444 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 9 |
Hb_001366_290 |
0.0888646923 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000418_170 |
0.0889267492 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 11 |
Hb_004218_140 |
0.0918151204 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 12 |
Hb_001157_050 |
0.0965346309 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 13 |
Hb_001488_420 |
0.0984711148 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 14 |
Hb_008304_010 |
0.1014132566 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 15 |
Hb_128196_010 |
0.1044927609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_039075_020 |
0.1061883601 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 17 |
Hb_000120_340 |
0.1070648266 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 18 |
Hb_026240_020 |
0.1071293937 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 19 |
Hb_004592_010 |
0.1100297129 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 20 |
Hb_004117_280 |
0.1105231872 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |