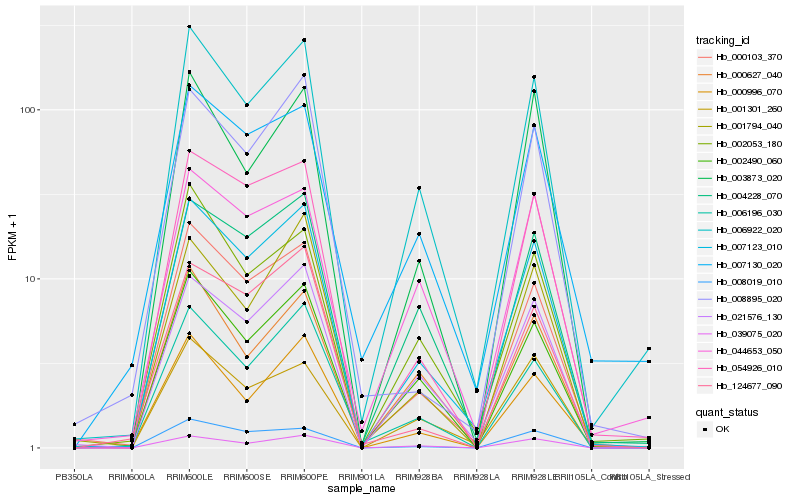
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007123_010 |
0.0 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 2 |
Hb_006922_020 |
0.0588211388 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 3 |
Hb_000103_370 |
0.0648423866 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 4 |
Hb_021576_130 |
0.0802830562 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004228_070 |
0.0866779987 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 6 |
Hb_054926_010 |
0.0875187603 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 7 |
Hb_002490_060 |
0.0897688071 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 8 |
Hb_006196_030 |
0.0941033072 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 9 |
Hb_039075_020 |
0.0954563018 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 10 |
Hb_000996_070 |
0.1025167077 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_008019_010 |
0.1041083387 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 12 |
Hb_007130_020 |
0.110521287 |
- |
- |
- |
| 13 |
Hb_002053_180 |
0.111483425 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 14 |
Hb_001794_040 |
0.1116965828 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 15 |
Hb_124677_090 |
0.1133570144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008895_020 |
0.1154249591 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 17 |
Hb_003873_020 |
0.115710266 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 18 |
Hb_044653_050 |
0.1169673208 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001301_260 |
0.1182800278 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000627_040 |
0.1187228156 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |