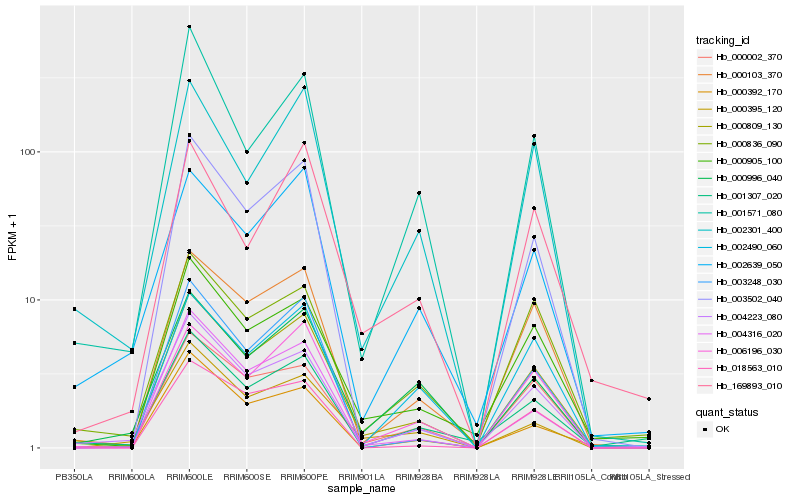
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_130 |
0.0 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 2 |
Hb_004223_080 |
0.0932243751 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000103_370 |
0.0953710804 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 4 |
Hb_006196_030 |
0.1029441646 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 5 |
Hb_000905_100 |
0.1064961573 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000002_370 |
0.1089564973 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 7 |
Hb_002301_400 |
0.1149852834 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 8 |
Hb_003248_030 |
0.1167000068 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_001307_020 |
0.1215466119 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 10 |
Hb_000996_040 |
0.1233040109 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 11 |
Hb_001571_080 |
0.123306415 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 12 |
Hb_000392_170 |
0.1239874723 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 13 |
Hb_002639_050 |
0.124205057 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 14 |
Hb_000836_090 |
0.1250427293 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 15 |
Hb_003502_040 |
0.1269578708 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 16 |
Hb_004316_020 |
0.1291123951 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 17 |
Hb_002490_060 |
0.1309697963 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 18 |
Hb_018563_010 |
0.1316443479 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 19 |
Hb_000395_120 |
0.1322431231 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 20 |
Hb_169893_010 |
0.1325054423 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |