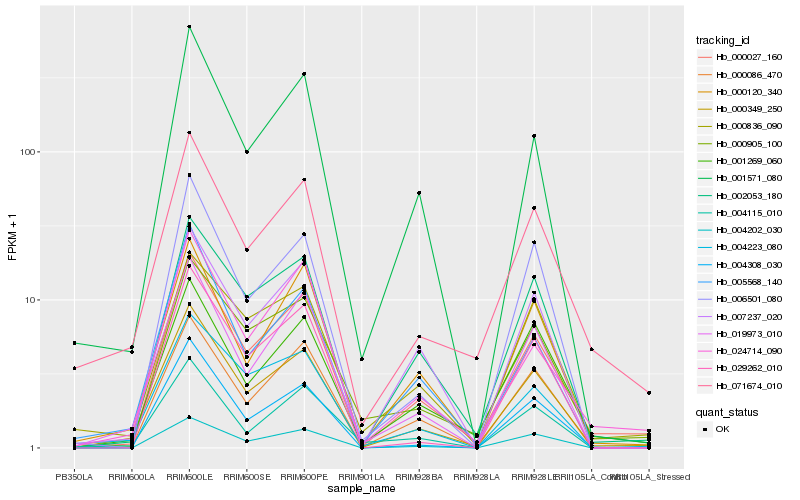
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000349_250 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 2 |
Hb_000027_160 |
0.075684372 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 3 |
Hb_019973_010 |
0.087624905 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002053_180 |
0.1031180569 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_071674_010 |
0.1044010539 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 6 |
Hb_004223_080 |
0.1079789539 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_001571_080 |
0.1087403884 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 8 |
Hb_004202_030 |
0.1099553246 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 9 |
Hb_006501_080 |
0.1100600398 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 10 |
Hb_000120_340 |
0.1116839322 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 11 |
Hb_004308_030 |
0.1119135694 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 12 |
Hb_001269_060 |
0.1122899223 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000086_470 |
0.1124846143 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 14 |
Hb_007237_020 |
0.1154662691 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 15 |
Hb_005568_140 |
0.1199075419 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_029262_010 |
0.1247118709 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 17 |
Hb_000905_100 |
0.1263426885 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000836_090 |
0.1299147207 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 19 |
Hb_004115_010 |
0.1300124898 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 20 |
Hb_024714_090 |
0.1300234881 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |