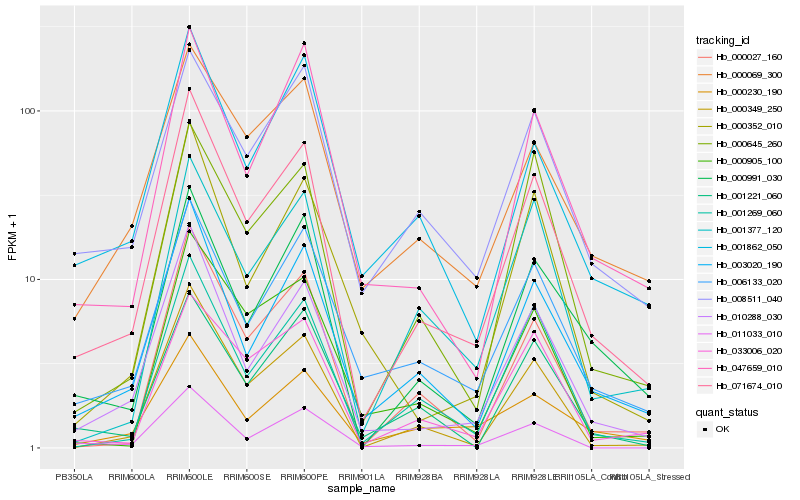
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071674_010 |
0.0 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 2 |
Hb_010288_030 |
0.0801940403 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 3 |
Hb_003020_190 |
0.0939729931 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 4 |
Hb_000349_250 |
0.1044010539 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 5 |
Hb_000027_160 |
0.1159128472 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000991_030 |
0.1167039692 |
- |
- |
PREDICTED: uncharacterized protein LOC105122436 [Populus euphratica] |
| 7 |
Hb_001269_060 |
0.1171533302 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006133_020 |
0.1221124434 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 9 |
Hb_011033_010 |
0.1269941775 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_047659_010 |
0.1320400606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_033006_020 |
0.1320515039 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 12 |
Hb_001862_050 |
0.1329039399 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 13 |
Hb_000069_300 |
0.1337643775 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000230_190 |
0.1357724556 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008511_040 |
0.1365395404 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_000645_260 |
0.1377323243 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 17 |
Hb_001377_120 |
0.1378234382 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 18 |
Hb_000352_010 |
0.1391350516 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000905_100 |
0.1393798623 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001221_060 |
0.1406950506 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |