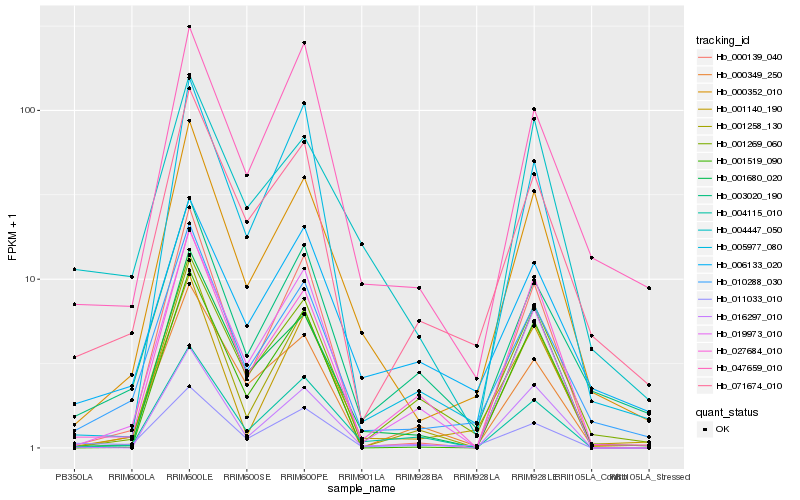
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_010 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 2 |
Hb_010288_030 |
0.0955272419 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 3 |
Hb_001680_020 |
0.1272282385 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 4 |
Hb_001140_190 |
0.1315032568 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 5 |
Hb_071674_010 |
0.1391350516 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 6 |
Hb_004115_010 |
0.139391003 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 7 |
Hb_000139_040 |
0.1442286446 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 8 |
Hb_001269_060 |
0.1452901516 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003020_190 |
0.1453710932 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 10 |
Hb_047659_010 |
0.1482478434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001519_090 |
0.1487291314 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 12 |
Hb_016297_010 |
0.1490435076 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 13 |
Hb_011033_010 |
0.1496612495 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_005977_080 |
0.1522666847 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 15 |
Hb_027684_010 |
0.1530364325 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 16 |
Hb_004447_050 |
0.1532372111 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_019973_010 |
0.154152007 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000349_250 |
0.1547407588 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 19 |
Hb_006133_020 |
0.1560102497 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 20 |
Hb_001258_130 |
0.1561951395 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |