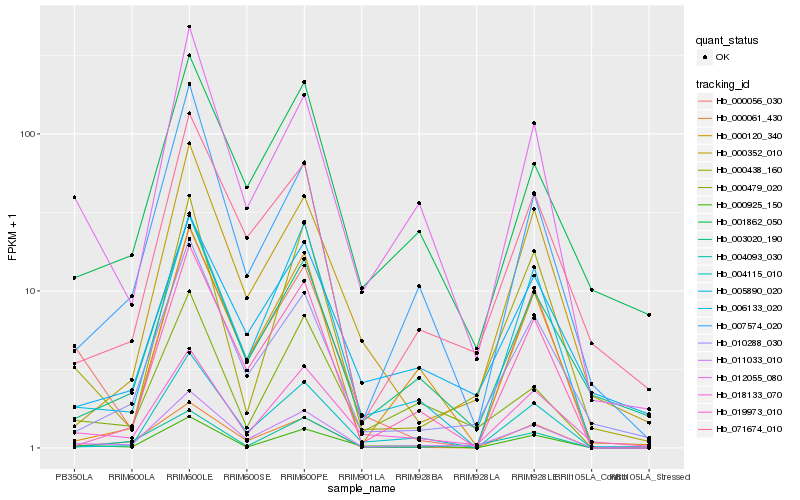
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011033_010 |
0.0 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_010288_030 |
0.1177756663 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 3 |
Hb_071674_010 |
0.1269941775 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 4 |
Hb_000061_430 |
0.1276085727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005890_020 |
0.1296131059 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 6 |
Hb_012055_080 |
0.1329316674 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 7 |
Hb_018133_070 |
0.1361561244 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 8 |
Hb_019973_010 |
0.1361961578 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004115_010 |
0.1364570406 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 10 |
Hb_000056_030 |
0.1413099421 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g14390 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003020_190 |
0.1450042238 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 12 |
Hb_000925_150 |
0.1494534207 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 13 |
Hb_000352_010 |
0.1496612495 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000438_160 |
0.1524051713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006133_020 |
0.1535724139 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 16 |
Hb_001862_050 |
0.1536128753 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 17 |
Hb_004093_030 |
0.1556690614 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 18 |
Hb_000120_340 |
0.1557029816 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 19 |
Hb_000479_020 |
0.1564557024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007574_020 |
0.1573225222 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |