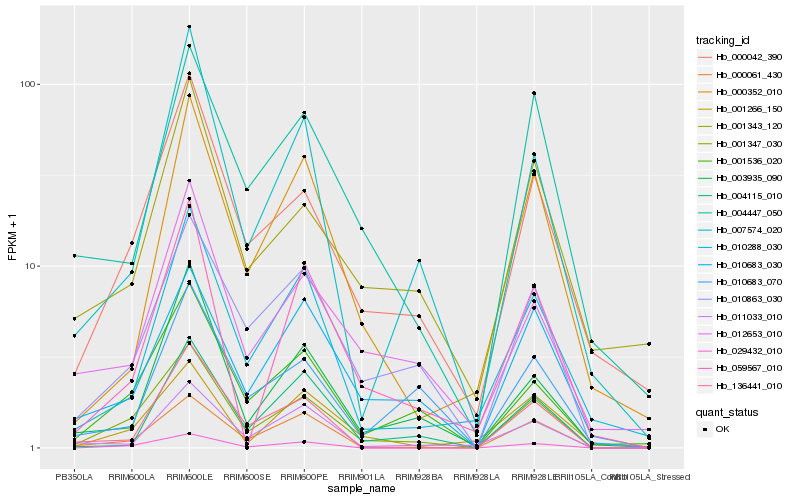
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001347_030 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_000042_390 |
0.1286490863 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 3 |
Hb_001536_020 |
0.1314832996 |
transcription factor |
TF Family: GRF |
PREDICTED: uncharacterized protein LOC105631006 [Jatropha curcas] |
| 4 |
Hb_010863_030 |
0.1444082972 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 5 |
Hb_000061_430 |
0.1462859299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_029432_010 |
0.1477923576 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_007574_020 |
0.171312086 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 8 |
Hb_010683_030 |
0.1724990871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010288_030 |
0.1767557708 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 10 |
Hb_003935_090 |
0.1808544178 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 11 |
Hb_012653_010 |
0.1815461225 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 12 |
Hb_136441_010 |
0.1855175196 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 13 |
Hb_004447_050 |
0.1871539826 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001343_120 |
0.1891802473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011033_010 |
0.1908330171 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 16 |
Hb_059567_010 |
0.1929192399 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 17 |
Hb_001266_150 |
0.1942997392 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 18 |
Hb_010683_070 |
0.196820349 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000352_010 |
0.1991531961 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004115_010 |
0.2002477016 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |