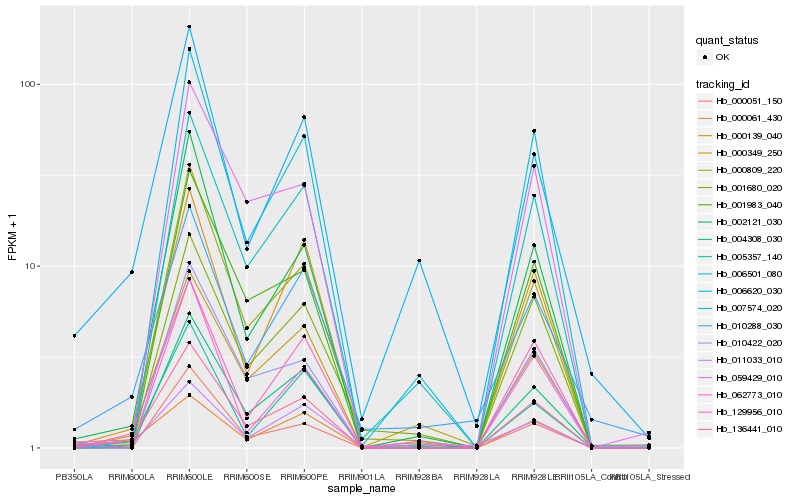
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136441_010 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 2 |
Hb_002121_030 |
0.1328628617 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 3 |
Hb_000809_220 |
0.1345862148 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 4 |
Hb_000061_430 |
0.1357096277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_062773_010 |
0.1438479529 |
- |
- |
beta glucosidase [Manihot esculenta] |
| 6 |
Hb_000139_040 |
0.1441335821 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 7 |
Hb_129956_010 |
0.1452325977 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 [Eucalyptus grandis] |
| 8 |
Hb_001983_040 |
0.1464674648 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 9 |
Hb_006620_030 |
0.1527458743 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 10 |
Hb_004308_030 |
0.1536518392 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 11 |
Hb_006501_080 |
0.154073789 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 12 |
Hb_010288_030 |
0.1556193925 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 13 |
Hb_005357_140 |
0.1573309714 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 14 |
Hb_010422_020 |
0.1576697161 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 15 |
Hb_007574_020 |
0.1580486632 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 16 |
Hb_059429_010 |
0.1581401353 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 17 |
Hb_001680_020 |
0.1584649937 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 18 |
Hb_011033_010 |
0.1598863667 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 19 |
Hb_000349_250 |
0.1601079665 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 20 |
Hb_000051_150 |
0.1606852921 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |