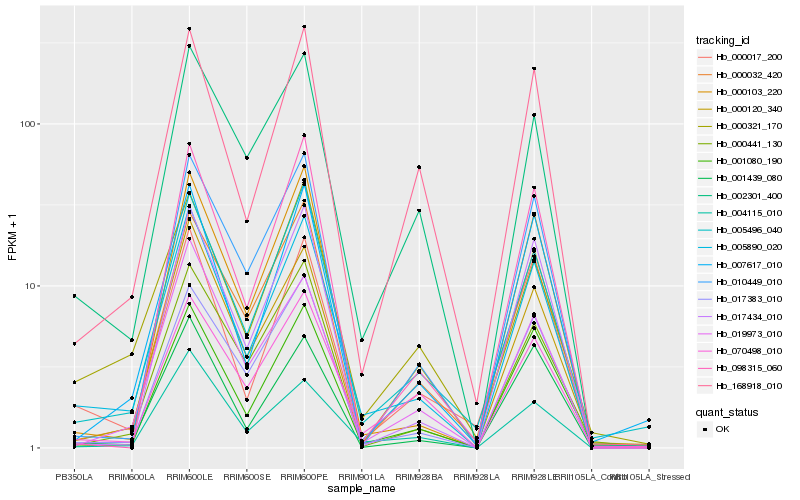
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005890_020 |
0.0 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 2 |
Hb_005496_040 |
0.0951391583 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 3 |
Hb_001080_190 |
0.1075563197 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002301_400 |
0.1088188796 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 5 |
Hb_001439_080 |
0.1106660935 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 6 |
Hb_017383_010 |
0.1109772409 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 7 |
Hb_000103_220 |
0.1116403786 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 8 |
Hb_000017_200 |
0.1134282014 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 9 |
Hb_004115_010 |
0.1140656406 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 10 |
Hb_007617_010 |
0.11466584 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000032_420 |
0.116995673 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 12 |
Hb_000321_170 |
0.1176420325 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 13 |
Hb_000120_340 |
0.1177624044 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 14 |
Hb_000441_130 |
0.118050769 |
- |
- |
PREDICTED: homeobox-leucine zipper protein HOX32 [Vitis vinifera] |
| 15 |
Hb_019973_010 |
0.1181218726 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 16 |
Hb_168918_010 |
0.1201264535 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 17 |
Hb_010449_010 |
0.1205941819 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 18 |
Hb_070498_010 |
0.120627557 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 19 |
Hb_017434_010 |
0.1224872006 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 20 |
Hb_098315_060 |
0.1252518774 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |