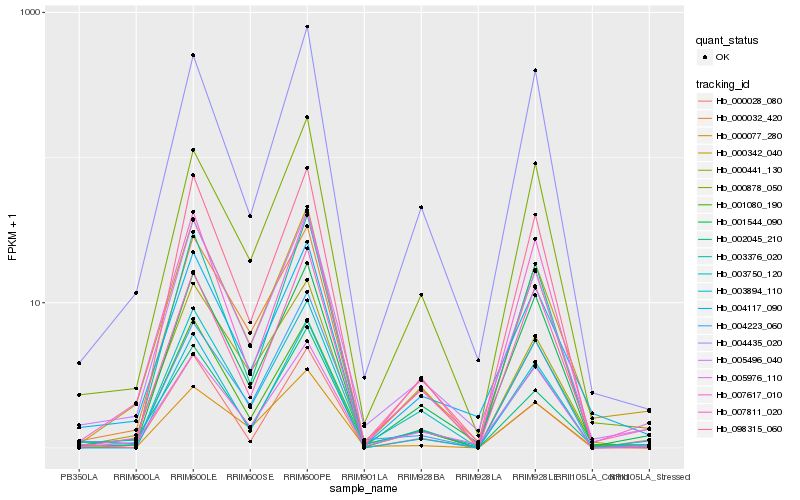
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000342_040 |
0.0 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 2 |
Hb_005496_040 |
0.076730076 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 3 |
Hb_002045_210 |
0.0921507801 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 4 |
Hb_004117_090 |
0.0942545104 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 5 |
Hb_000441_130 |
0.0950347527 |
- |
- |
PREDICTED: homeobox-leucine zipper protein HOX32 [Vitis vinifera] |
| 6 |
Hb_000032_420 |
0.0956328902 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 7 |
Hb_001544_090 |
0.0984956476 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 8 |
Hb_007617_010 |
0.1017095054 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003750_120 |
0.1022445004 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 10 |
Hb_001080_190 |
0.104020468 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007811_020 |
0.1096343909 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 12 |
Hb_000878_050 |
0.1105471492 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 13 |
Hb_003894_110 |
0.1117850332 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 14 |
Hb_098315_060 |
0.1146344601 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 15 |
Hb_005976_110 |
0.1152118684 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 16 |
Hb_004435_020 |
0.1156837086 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 17 |
Hb_003376_020 |
0.1159431477 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000077_280 |
0.1164237983 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000028_080 |
0.1165009997 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 20 |
Hb_004223_060 |
0.1173659313 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |