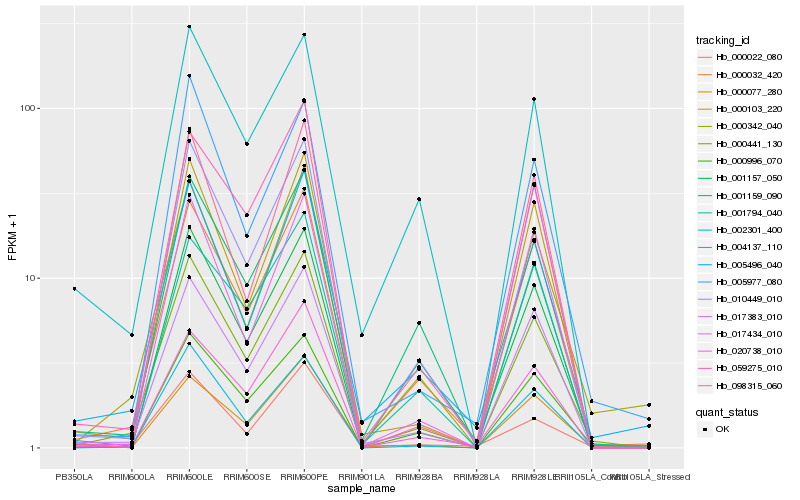
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000441_130 |
0.0 |
- |
- |
PREDICTED: homeobox-leucine zipper protein HOX32 [Vitis vinifera] |
| 2 |
Hb_000996_070 |
0.0765758293 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000032_420 |
0.0789738893 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 4 |
Hb_017383_010 |
0.0824539144 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 5 |
Hb_010449_010 |
0.085035087 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 6 |
Hb_004137_110 |
0.0876962491 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |
| 7 |
Hb_000077_280 |
0.0945019218 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000342_040 |
0.0950347527 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 9 |
Hb_005496_040 |
0.0983722073 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 10 |
Hb_000103_220 |
0.0987056523 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 11 |
Hb_020738_010 |
0.0990431573 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 12 |
Hb_005977_080 |
0.1073588695 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 13 |
Hb_059275_010 |
0.1082998167 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 14 |
Hb_000022_080 |
0.1083740114 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 15 |
Hb_098315_060 |
0.1131107166 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 16 |
Hb_002301_400 |
0.1152304948 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 17 |
Hb_001157_050 |
0.1156020437 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 18 |
Hb_001159_090 |
0.1159272496 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 19 |
Hb_017434_010 |
0.1161918023 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 20 |
Hb_001794_040 |
0.1176657511 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |