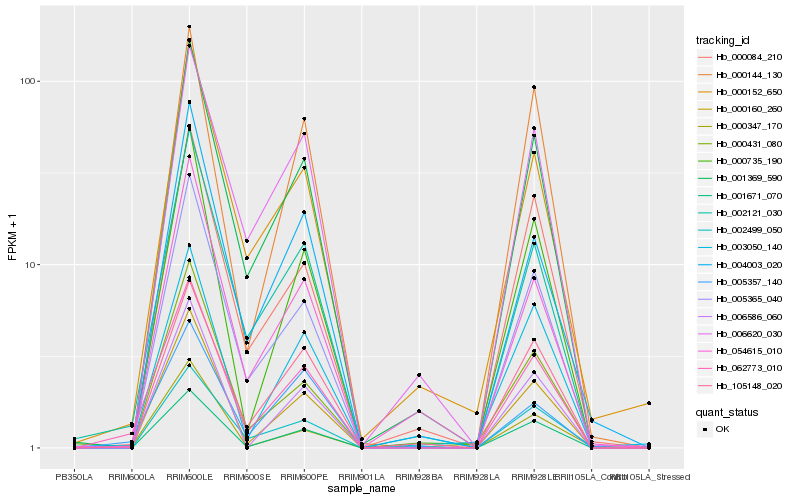
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062773_010 |
0.0 |
- |
- |
beta glucosidase [Manihot esculenta] |
| 2 |
Hb_002121_030 |
0.094377804 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 3 |
Hb_000160_260 |
0.1034810254 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 4 |
Hb_001369_590 |
0.1035525508 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 5 |
Hb_005365_040 |
0.1054990837 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 6 |
Hb_001671_070 |
0.1099153735 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_105148_020 |
0.1154887725 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 8 |
Hb_000735_190 |
0.1214831434 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 9 |
Hb_000152_650 |
0.126316113 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_000084_210 |
0.1284367115 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 11 |
Hb_000144_130 |
0.1312481268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005357_140 |
0.1315808052 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 13 |
Hb_003050_140 |
0.1318517499 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 14 |
Hb_000347_170 |
0.1319392834 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 15 |
Hb_054615_010 |
0.132493973 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 16 |
Hb_004003_020 |
0.1329748925 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_006586_060 |
0.1334477371 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 18 |
Hb_002499_050 |
0.1345761824 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000431_080 |
0.1346453042 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 20 |
Hb_006620_030 |
0.1347207163 |
- |
- |
nitrate reductase, putative [Ricinus communis] |