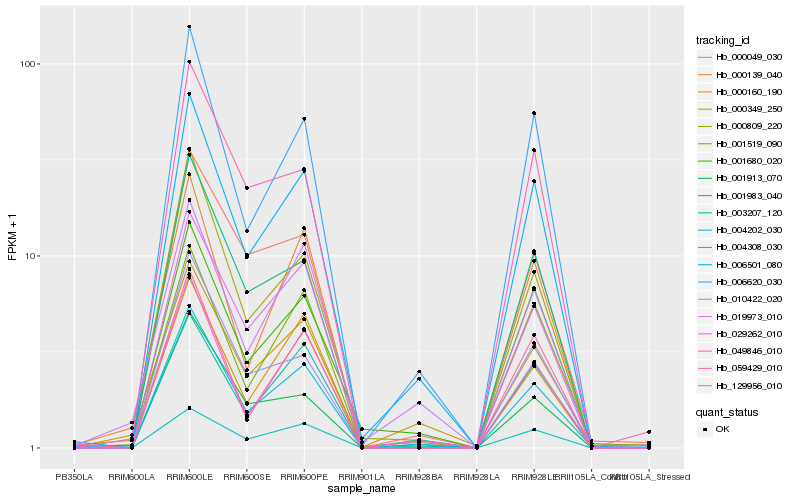
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004308_030 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 2 |
Hb_006501_080 |
0.0624810524 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 3 |
Hb_029262_010 |
0.0769679889 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 4 |
Hb_006620_030 |
0.078386858 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 5 |
Hb_000809_220 |
0.0890701736 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 6 |
Hb_049846_010 |
0.0961699966 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 7 |
Hb_004202_030 |
0.1017851088 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 8 |
Hb_001983_040 |
0.1032420192 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 9 |
Hb_000160_190 |
0.1046244345 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 10 |
Hb_003207_120 |
0.1091025537 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 11 |
Hb_000049_030 |
0.1096403495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001913_070 |
0.1106362754 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 13 |
Hb_000349_250 |
0.1119135694 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 14 |
Hb_001680_020 |
0.1119793795 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 15 |
Hb_001519_090 |
0.1133565888 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 16 |
Hb_000139_040 |
0.1138182117 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 17 |
Hb_129956_010 |
0.117101228 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 [Eucalyptus grandis] |
| 18 |
Hb_010422_020 |
0.1206241727 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 19 |
Hb_019973_010 |
0.1214897871 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 20 |
Hb_059429_010 |
0.1222743626 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |