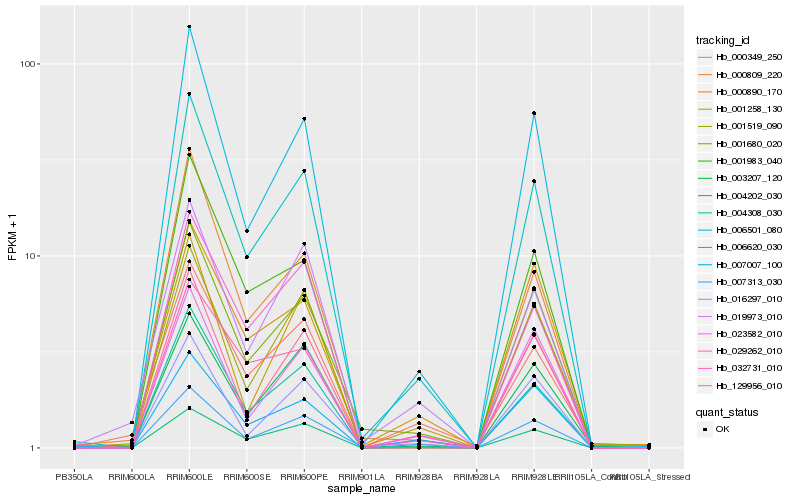
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006501_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 2 |
Hb_006620_030 |
0.0618777062 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 3 |
Hb_004308_030 |
0.0624810524 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 4 |
Hb_001680_020 |
0.0704496142 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 5 |
Hb_004202_030 |
0.0759213031 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 6 |
Hb_029262_010 |
0.0929314852 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 7 |
Hb_003207_120 |
0.0946532921 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 8 |
Hb_007007_100 |
0.1037275139 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 9 |
Hb_016297_010 |
0.1042174539 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 10 |
Hb_000809_220 |
0.1049587535 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 11 |
Hb_032731_010 |
0.1052936937 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 12 |
Hb_023582_010 |
0.1068294875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_129956_010 |
0.1080428152 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 [Eucalyptus grandis] |
| 14 |
Hb_001983_040 |
0.1082161707 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 15 |
Hb_019973_010 |
0.1089491114 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007313_030 |
0.1092801006 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 17 |
Hb_001258_130 |
0.1095695651 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 18 |
Hb_000349_250 |
0.1100600398 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 19 |
Hb_000890_170 |
0.111271108 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 20 |
Hb_001519_090 |
0.1113014649 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |