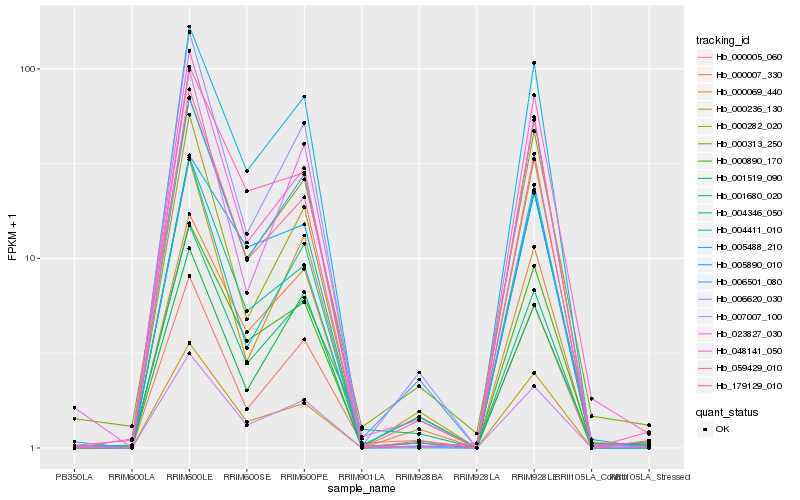
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007007_100 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 2 |
Hb_005488_210 |
0.061636238 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 3 |
Hb_000007_330 |
0.0729585801 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 4 |
Hb_000890_170 |
0.0900239699 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 5 |
Hb_000313_250 |
0.0920007483 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 6 |
Hb_000236_130 |
0.0943747469 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 7 |
Hb_006620_030 |
0.1000667998 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 8 |
Hb_023827_030 |
0.1003457617 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_001680_020 |
0.101611171 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 10 |
Hb_000282_020 |
0.1024869594 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004346_050 |
0.1029826581 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 12 |
Hb_006501_080 |
0.1037275139 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 13 |
Hb_005890_010 |
0.1045387298 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 14 |
Hb_179129_010 |
0.1047336976 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 15 |
Hb_004411_010 |
0.1063049588 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 16 |
Hb_001519_090 |
0.1063595483 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 17 |
Hb_059429_010 |
0.1080396287 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 18 |
Hb_000005_060 |
0.1083845931 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 19 |
Hb_000069_440 |
0.1084628455 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 20 |
Hb_048141_050 |
0.1088781784 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |