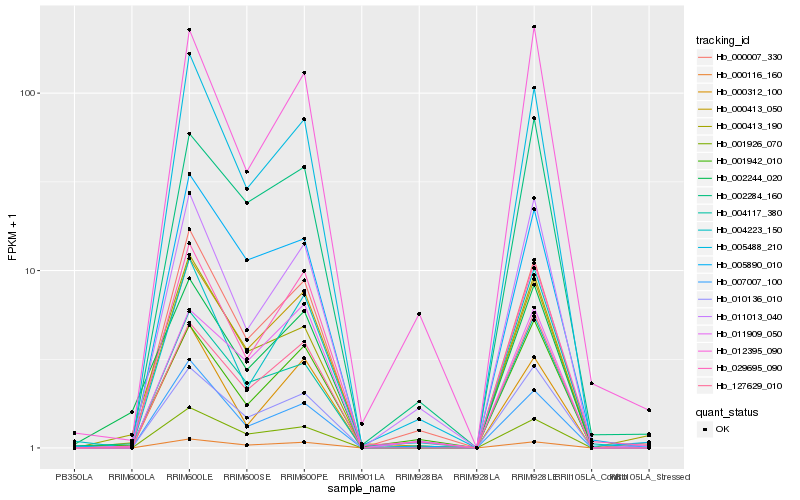
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000413_050 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 2 |
Hb_029695_090 |
0.0673432925 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 3 |
Hb_000116_160 |
0.0697840584 |
transcription factor |
TF Family: RWP-RK |
hypothetical protein JCGZ_22391 [Jatropha curcas] |
| 4 |
Hb_000007_330 |
0.0774459929 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 5 |
Hb_005488_210 |
0.0827026982 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 6 |
Hb_127629_010 |
0.0999955627 |
- |
- |
hypothetical protein CICLE_v10005216mg [Citrus clementina] |
| 7 |
Hb_005890_010 |
0.1026882537 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 8 |
Hb_001942_010 |
0.1085909409 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 9 |
Hb_001926_070 |
0.1088143789 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 10 |
Hb_007007_100 |
0.1116756217 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 11 |
Hb_004117_380 |
0.1118048467 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 12 |
Hb_000413_190 |
0.1155855206 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 13 |
Hb_002284_160 |
0.115965901 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_012395_090 |
0.1164724177 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004223_150 |
0.1167742499 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 16 |
Hb_010136_010 |
0.1175589067 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 17 |
Hb_002244_020 |
0.1178872489 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 18 |
Hb_011909_050 |
0.1181574069 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 19 |
Hb_011013_040 |
0.1182221609 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 20 |
Hb_000312_100 |
0.1192373173 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |