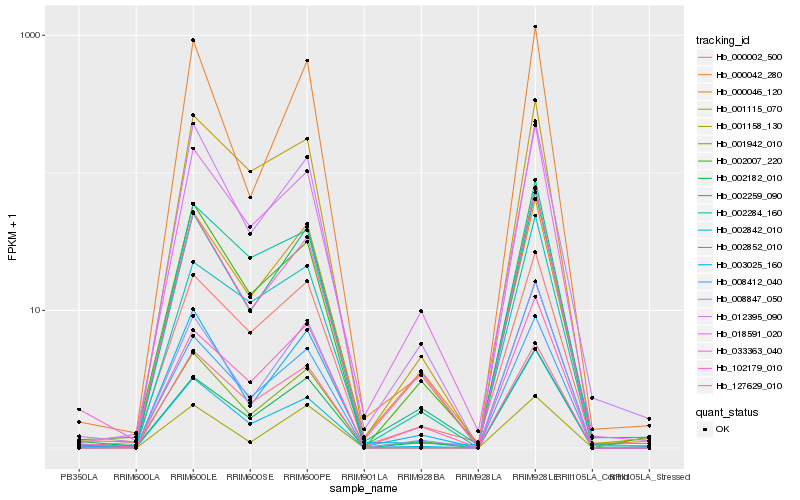
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008412_040 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_127629_010 |
0.062641214 |
- |
- |
hypothetical protein CICLE_v10005216mg [Citrus clementina] |
| 3 |
Hb_002259_090 |
0.0867661614 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 4 |
Hb_000002_500 |
0.0881822523 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001942_010 |
0.0930689852 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 6 |
Hb_001115_070 |
0.0941559519 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |
| 7 |
Hb_008847_050 |
0.0950503261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002852_010 |
0.0960740848 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_002007_220 |
0.100445743 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002842_010 |
0.1022534515 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 11 |
Hb_002182_010 |
0.1032867939 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Jatropha curcas] |
| 12 |
Hb_000046_120 |
0.1033747856 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_033363_040 |
0.1048118325 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 14 |
Hb_002284_160 |
0.104882934 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_012395_090 |
0.105025273 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 16 |
Hb_018591_020 |
0.1057579181 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000042_280 |
0.1068055736 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 18 |
Hb_102179_010 |
0.1097433465 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_003025_160 |
0.1103067669 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 20 |
Hb_001158_130 |
0.1109984077 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |