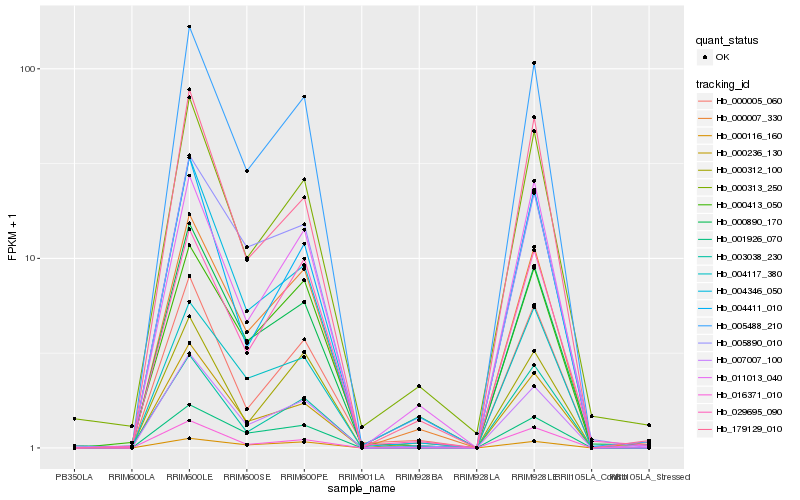
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005488_210 |
0.0 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 2 |
Hb_000007_330 |
0.0520471377 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 3 |
Hb_007007_100 |
0.061636238 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 4 |
Hb_000413_050 |
0.0827026982 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005890_010 |
0.0848350075 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 6 |
Hb_000236_130 |
0.0873285585 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 7 |
Hb_004346_050 |
0.0874222975 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 8 |
Hb_004117_380 |
0.0885345461 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 9 |
Hb_000005_060 |
0.0888859298 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 10 |
Hb_029695_090 |
0.0897135911 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 11 |
Hb_004411_010 |
0.090783384 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 12 |
Hb_179129_010 |
0.0910635296 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 13 |
Hb_000116_160 |
0.0918455523 |
transcription factor |
TF Family: RWP-RK |
hypothetical protein JCGZ_22391 [Jatropha curcas] |
| 14 |
Hb_016371_010 |
0.0919986251 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 15 |
Hb_000890_170 |
0.0932021312 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 16 |
Hb_000312_100 |
0.0946343874 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 17 |
Hb_000313_250 |
0.0947043821 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 18 |
Hb_003038_230 |
0.0955942422 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 19 |
Hb_001926_070 |
0.0955948167 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 20 |
Hb_011013_040 |
0.0958240511 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |