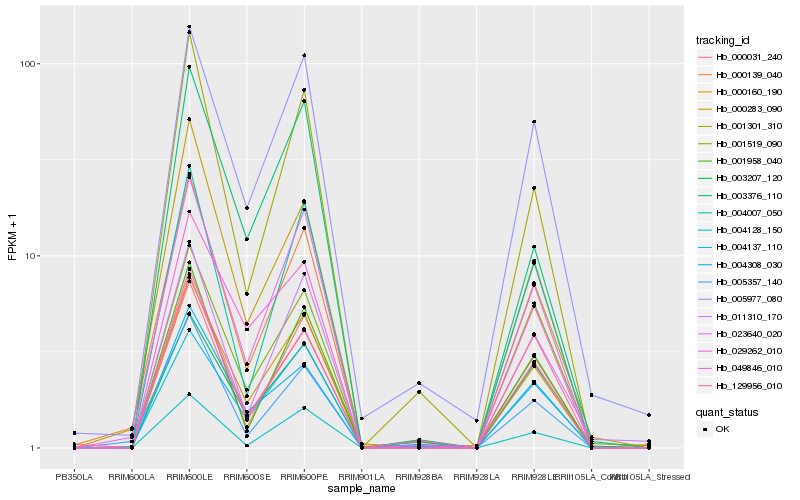
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_190 |
0.0 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 2 |
Hb_023640_020 |
0.0610525565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_049846_010 |
0.0721674085 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 4 |
Hb_005977_080 |
0.0853374676 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 5 |
Hb_029262_010 |
0.0873593106 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 6 |
Hb_004137_110 |
0.0914032794 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |
| 7 |
Hb_004128_150 |
0.095706238 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 8 |
Hb_004007_050 |
0.0968491869 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 9 |
Hb_000139_040 |
0.0995369279 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 10 |
Hb_011310_170 |
0.1008443373 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_004308_030 |
0.1046244345 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 12 |
Hb_000031_240 |
0.1066331589 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 13 |
Hb_003376_110 |
0.1081938038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001301_310 |
0.1085056215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001958_040 |
0.1104228971 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 16 |
Hb_001519_090 |
0.1126418508 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 17 |
Hb_003207_120 |
0.1218098701 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 18 |
Hb_129956_010 |
0.1247677263 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 [Eucalyptus grandis] |
| 19 |
Hb_000283_090 |
0.1268705543 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_005357_140 |
0.1281917836 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |