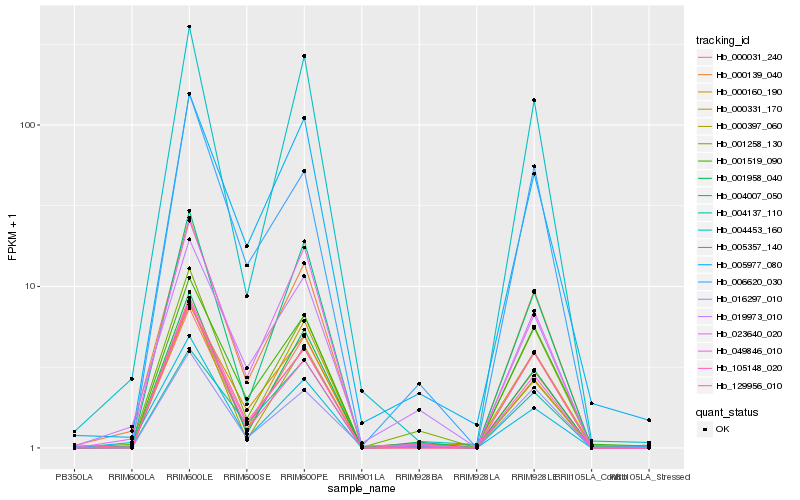
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_040 |
0.0 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 2 |
Hb_049846_010 |
0.0741657052 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 3 |
Hb_001258_130 |
0.0761241704 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 4 |
Hb_129956_010 |
0.0823257815 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 [Eucalyptus grandis] |
| 5 |
Hb_023640_020 |
0.0845483507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000031_240 |
0.0859297664 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 7 |
Hb_005357_140 |
0.0884416773 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 8 |
Hb_004007_050 |
0.0889309557 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 9 |
Hb_001519_090 |
0.0904891505 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 10 |
Hb_004453_160 |
0.0926776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001958_040 |
0.0935451099 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 12 |
Hb_000397_060 |
0.0947106269 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 13 |
Hb_105148_020 |
0.0973976636 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 14 |
Hb_000160_190 |
0.0995369279 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 15 |
Hb_019973_010 |
0.1040855786 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005977_080 |
0.1048336949 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 17 |
Hb_006620_030 |
0.1058563572 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 18 |
Hb_004137_110 |
0.1063545172 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |
| 19 |
Hb_016297_010 |
0.1079034677 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 20 |
Hb_000331_170 |
0.1088166231 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |